Figure 5. Super-enhancer sequences and neighboring conventional enhancers or coding genes express RNA exosome substrate antisense RNAs.
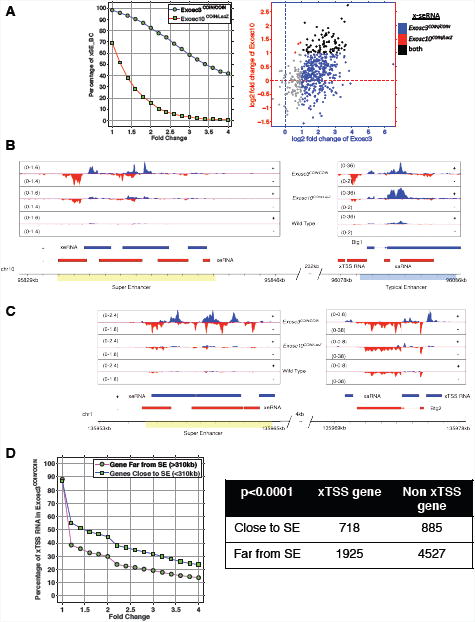
(A) Left panel: expression of super-enhancer RNAs at 200 annotated super-enhancers within Exosc3 (Exosc3COIN/COIN) and Exosc10 (Exosc10COIN/LacZ) exotomes. X-axis indicates the cutoff of fold change, and y-axis indicates the fraction of super-enhancers with higher expression (given x-axis) compared with wild type. Right panel: expression of Exosc3 (blue), Exosc10 (red), and overlapping (black) x-seRNAs in B cells. (B) A super-enhancer resident in chromosome 10 (SEChr10) and neighboring conventional enhancer element resident at the Btg1 locus express sense (blue) and antisense (red) x-eRNAs. (C) A super-enhancer resident in chromosome 1 (SEChr1) and neighboring a conventional enhancer element resident at the Btg2 locus express sense (blue) and antisense (red) x-eRNAs. (D) Genome-wide correlation between proximity of super-enhancer location and antisense x-TSS-RNA expression at neighboring genes in B cells. See Figure S5.
