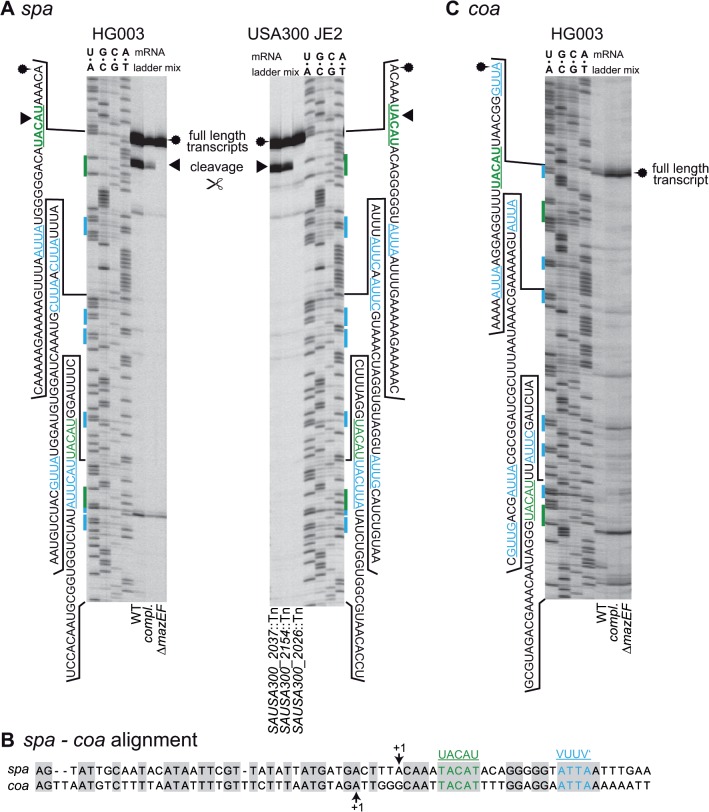
Fig 1. Cleavage of transcripts isolated from S. aureus.
(A) Cleavage of spa transcript in S. aureus HG003 and NARSA transposon mutant library derivatives of strain USA300 JE2. The spa transcript was cleaved in vivo at the first 5’ UACAU site (green) in HG003 and the complemented mazEF mutant. Cleavage did not occur at the VUUV’ sites (blue) and in the mazEF deletion mutant. Cleavage was also visible at the first UACAU site but not at VUUV’ in the transposon mutant strains NE565 (RNA helicase, SAUSA300_2037) and NE1535 (galactose-6-phosphate isomerase, SAUSA300_2154). The UACAU site is not cut in the mazF mutant strain NE1833 (SAUSA300_2026). RNA sequences of sequencing ladders are shown on the sides of the gels. Putative cleavage sites are marked in the sequence and sequencing ladders by green and blue text and bars. Cleavage is denoted by a black arrow head and the full length transcript is indicated by a fuzzy circle. (B) Alignment of the transcript and upstream region of spa and coa. The sequences of spa and coa possess conserved UACAU and VUUV’ sites. Identical bases are shaded in gray. +1 denotes TSP. (C) Test for MazF-dependent cleavage of coa transcript. Cleavage at UACAU or VUUV’ could not be observed in any of the examined strains despite high similarity to the spa transcript. WT = HG003 (pRAB11-gfpmut2), ΔmazEF = HG003ΔmazEF (pRAB11-gfpmut2), compl. = HG003ΔmazEF (pRAB11-P mazEF- mazEF).