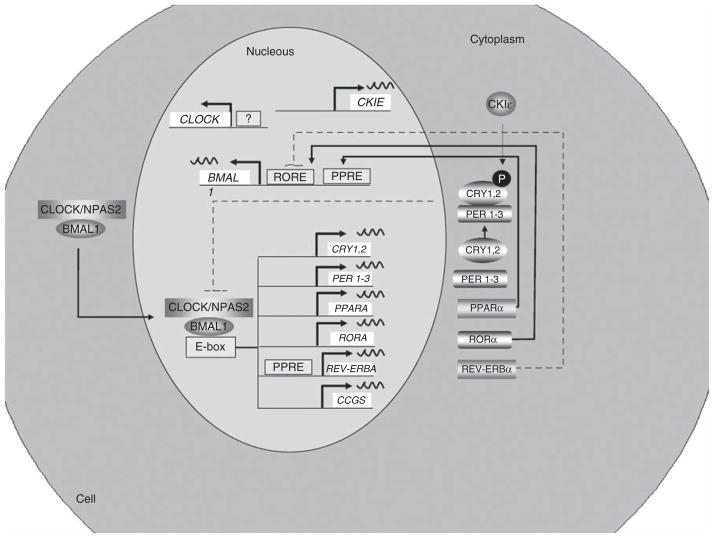
Figure 2.
Organization of the mammalian circadian intracellular oscillator. The cellular oscillator is composed of a positive (CLOCK and BMAL1) and a negative (PER1–3 and CRY1, 2) limb. CLOCK–BMAL1 heterodimeres, through binding to E-box elements, drive the transcription of several genes: Cry1, Cry2, Per1, Per2, Per3, Rev-Erbα, Rorα and multiple CCGs. After dimerization PERs and CRYs undergo nuclear translocation inhibiting CLOCK–BMAL1-mediated transcription. Once the levels of PERs and CRYs fall, the negative repression is lifted and CLOCK–BMAL1 binds again to the E-box. A secondary stabilizing loop is established by the negative, REV-ERBα, and positive, RORα, effect on Bmal1 transcription through their activity on RORE. In addition, the PPARα, a CCG, induces Bmal1 and Rev-Erbα transcription through its action on PPAR-response elements located in their respective promoters. The molecular circadian clock in linked to metabolism through several mechanisms. RORα and Rev-Erbα regulates lipid metabolism and adipogenesis. BMAL1, brain- and muscle ANRT-like protein-1; CCG, Clock-controlled gene; CLOCK, Circadian Locomotor Output Cycles Kaput; CRY, cryptochrome circadian protein; PER, Period Circadian Protein; PPAR, peroxisome proliferator-activated receptor; REV-ERBα, erythroblastosis virus-α; RORα, retinoic acid receptor-related orphan receptor-α.