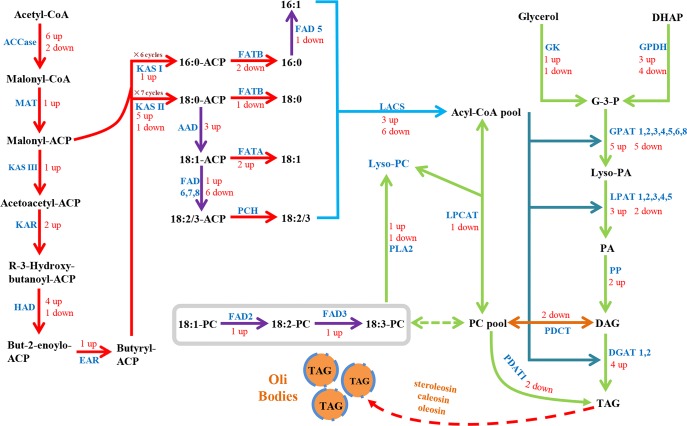
Fig 5. Overview of de novo fatty acid (FA) and triacylglycerol (TAG) biosynthesis pathways.
Indentified enzymes include: ACCase, aacetyl-CoA carboxylase carboxyl transferase (EC:6.4.1.2); MAT, Malonyl-CoA-ACP transacylase (EC:2.3.1.39); KAS, 3- oxoacyl ACP synthase (KASI, EC: 2.3.1.41; KASII, EC: 2.3.1.179; KAS III, EC: 2.3.1.180); KAR, 3-oxoacyl ACP reductase (EC:1.1.1.100); HAD, 3R-hydroxymyristoyl ACP dehydrase (EC:4.2.1.-); EAR, enoyl-ACP reductase I (EC:1.3.1.9); FATA/B, fatty acyl-ACP thioesterase A/B (EC:3.1.2.14 3.1.2.-); AAD, acyl-ACP desaturase (EC:1.14.19.2); PCH, palmitoyl-CoA hydrolase (EC:3.1.2.2); LACS, long-chain acyl-CoA synthetase (EC:6.2.1.3); FAD2/6, D12(ω6)-Desaturase (EC:1.14.19.-); FAD3/7/8, D15(ω3)-Desaturase (EC:1.14.19.-); GK, glycerol kinase (EC:2.7.1.30); ATS1/GPAT, glycerol-3-phosphate acyltransferase (EC:2.3.1.15); LPAT, lysophosphatidyl acyltransferase (EC:2.3.1.51); PP, phosphatidate phosphatase (EC:3.1.3.4); DGAT1, diacylglycerol O-acyltransferase 1 (EC:2.3.1.20); PDAT1, phospholipid: diacylglycerol acyltransferase 1 (EC:2.3.1.158); LPCAT, lysophosphatidylcholine acyltransferase (EC:2.3.1.23 2.3.1.67); PLA2, Phospholipase A2 (EC:3.1.1.4). Numbers indicated the numbers of each enzyme, and “up” meant up-regulation comparing 25DAP seeds with leaves, and “down” meant down-regulation. Lipid substrates are abbreviated: 16:0, palmitic acid; 18:0, stearic acid; 18:1, oleic acid; 18:2, linoleic acid.