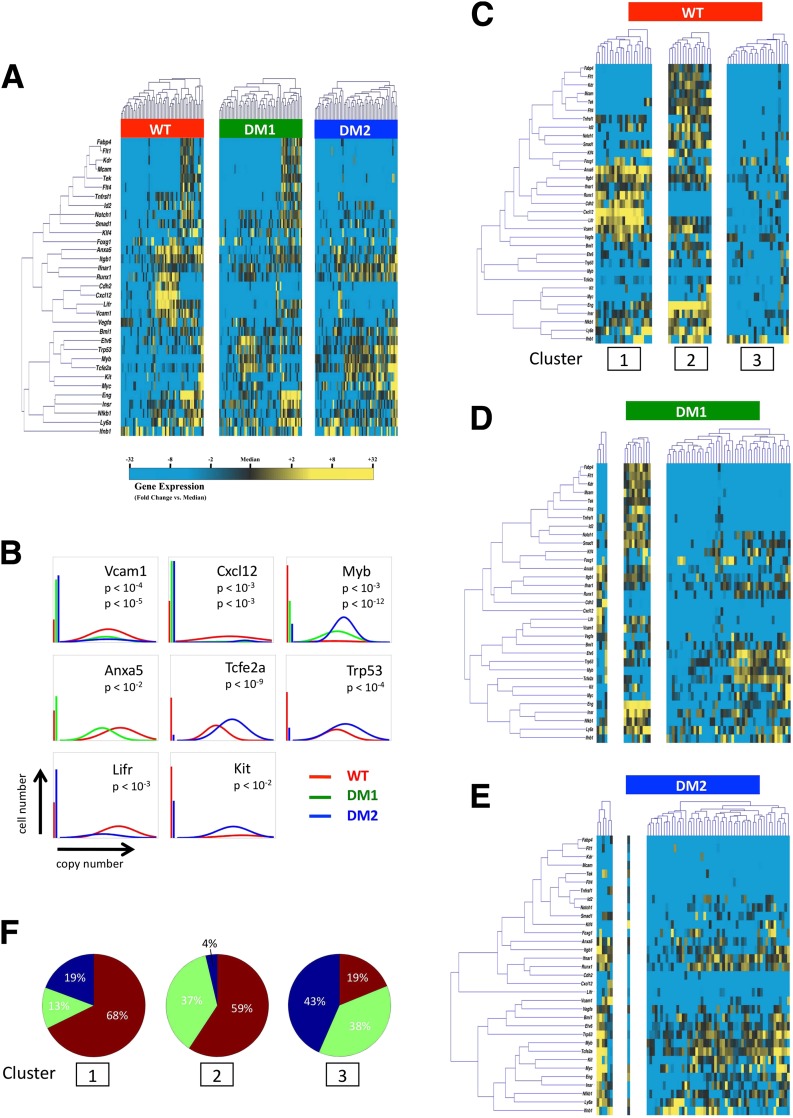
Figure 4.
Single-cell qPCR of Lin−/CD45−/Sca-1+ BM-MPCs. A: Hierarchical clustering of simultaneous gene expression for 60 individual cells from WT, STZ diabetic (DM1), and db/db (DM2) mice. Gene expression is presented as fold change from median on a color scale from yellow (high expression, 32-fold above median) to blue (low expression, 32-fold below median). See Supplementary Figs. 4 and 5 for complete data set. B: Differentially expressed genes between WT and diabetic (STZ [DM1] or db/db [DM2]) cells identified using nonparametric two-sample Kolmogorov-Smirnov testing. Eight genes exhibit significantly different (P < 0.01 after Bonferroni correction for multiple comparisons) distributions of single-cell expression between populations, illustrated here using median-centered Gaussian curve fits. The left bar for each panel represents the fraction of qPCR reactions that failed to amplify in each group. Curves and P values for each gene are shown only for those diabetic groups significantly different from WT cells. C: K-means clustering of WT cells, with k = 3 chosen using the gap statistic as described in Supplementary Fig. 6. D and E: Cells from STZ-induced (DM1) and db/db (DM2) animals were partitioned into subgroups according to the cluster centroids of the WT cells in C. F: Pie graphs representing the fraction of cells comprising each cluster from WT (red), STZ diabetic (green), and db/db (blue) animals.