Figure 3.
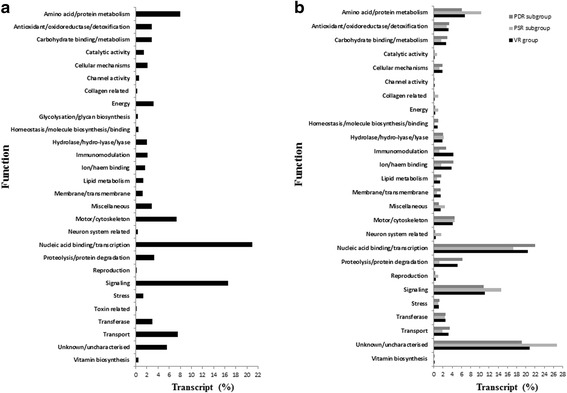
Relative abundances of the liver fluke transcripts in various biological functions based on manual inspection of protein motifs. The relative abundances (%) of all the functionally categorised transcripts within whole transcriptome (a), VR group and its PSR and PDR subgroups (b) in various functions are demonstrated. Nucleic acid binding/transcription and signaling mechanisms were highly pronounced functions of the whole transcriptome, with relative transcript abundances of 20.91% and 16.66%, respectively (Figure 3a). This pattern was similar within VR group except the relative transcript abundance of unknown mechanisms was higher within this group (20.47%), PSR subgroup (26.79%) and PDR subgroup (19.12 %). The transcript proportions of amino acid/protein metabolism, catalytic activity, energy, neuron system and signaling within PSR subgroup were higher (around 1.5-5 times) than those within PDR subgroup. The relative transcript ratios for immunomodulation, ion/haem binding and proteolysis/protein degradation functions were higher (about 2–5 times) in PDR subgroup in comparison with PSR subgroup.
