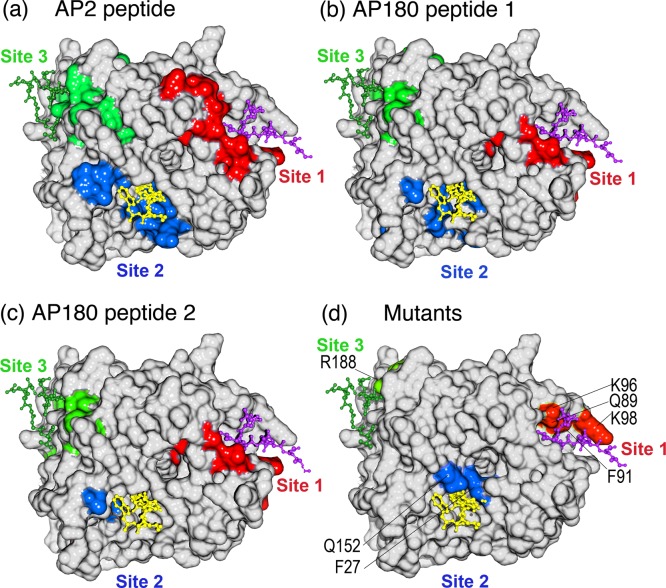
Figure 4.
Locations of shifted and broadened residues define three peptide binding sites on TD coincident with those previously defined by crystallography and mutational analyses. (a) Surface representation of TD structure in white with residue peaks that were either broadened or shifted by at least two standard deviations above the mean by the AP2 peptide in either red (site 1), blue (site 2), or green (site 3). (b) As in panel a, but for AP180 peptide 1. (c) As in panel a, but for AP180 peptide 2. (d) Mutations shown to affect binding to β-arrestin 2 (Q89, F91, K96, and K98),33 amphiphysin (F27 and Q152),4 and the β-arrestin 1 splice loop (R188)34 are highlighted on the TD structure in red, blue, and green, respectively. In all panels, a peptide from the β3 subunit of AP-3 [purple, Protein Data Bank (PDB) entry 1C9I], amphiphysin (yellow, PDB entry 1UTC), and the β-arrestin 1 splice loop (green, PDB entry 3GC3) are shown as they occur in the crystal structures of the respective TD–peptide complexes.