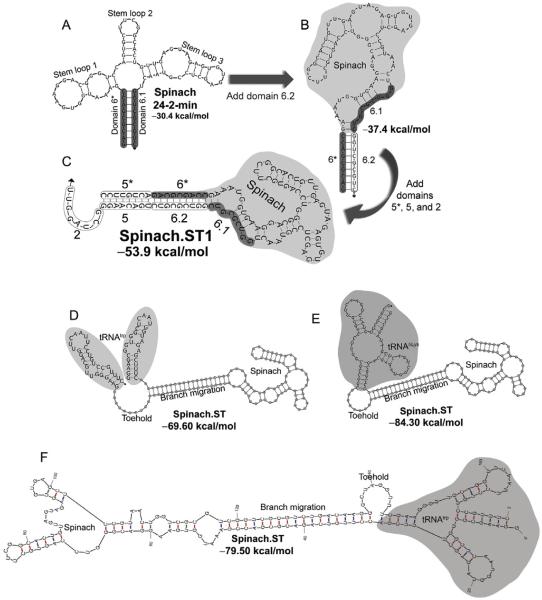
Figure 1.
Engineering the Spinach.ST molecular beacon. (A) The minimum free energy (MFE) structure of the minimized 80-mer RNA aptamer Spinach 24-2-min as predicted by NUPACK. Nine nucleotides at the 5′- and the 3′-ends of the aptamer (designated as domains 6* and 6.1, respectively) hybridize to form the basal stem (highlighted in gray). (B) The minimized Spinach aptamer was extended at its 3′-end with a duplicate domain 6.1 (designated as domain 6.2) and is predicted to fold into an alternate MFE conformation by NUPACK. Domain 6.2 hybridizes with domain 6* and disrupts the basal stem of the Spinach aptamer. (C) The NUPACK-predicted MFE conformation of Spinach when extended with an additional domain 5* at its 5′-end and with domains 2 and 5 at its 3′-end. The Spinach.ST1 sequence is depicted. The aptamer domains 6* and 6.1 that form the Spinach 24-2-min basal stem are unable to interact. The extended domains 5 and 6.2 (outlined) hybridize with complementary domains 5* and 6* to form a long stem that leaves the unpaired domain 2 free to act as a toehold. (D–F) The Spinach.ST molecular beacon is stabilized by flanking it with tRNA sequences that fold into hairpins. Structures of Saccharomyces cerevisiae tRNAtrp flanks (D) and human tRNALys flanks (E) as predicted by NUPACK are depicted. Structures of Saccharomyces cerevisiae tRNAtrp flanks generated by mFold are depicted in (E).