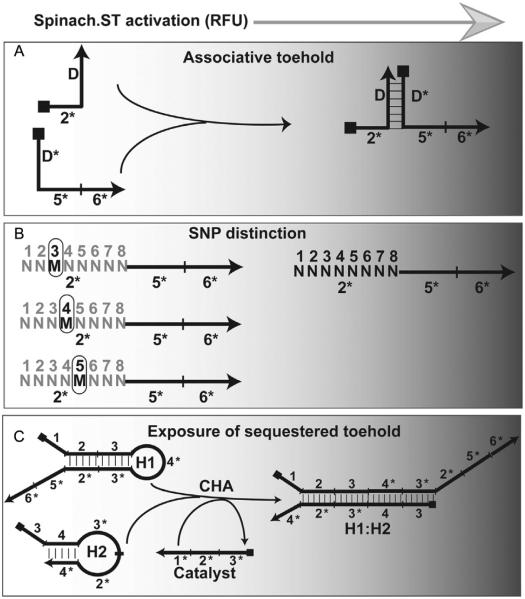
Figure 4.
Design options for Spinach.ST trigger sequences. (A) Generation of Spinach.ST trigger sequences by associative toehold activation. The trigger toehold domain 2* is placed at the 5′-end of one oligonucleotide, while the trigger branch migration domains 5* and 6* are placed at the 3′-end of a second oligonucleotide. Neither oligonucleotide alone is capable of activating Spinach.ST. However, when brought together by hybridization between the complementary domains D and D*, the toehold and branch migration domains activate Spinach.ST. (B) Presence of a single mismatched nucleotide (M) at position 3, 4, or 5 within the trigger toehold domain 2* significantly decreases the efficiency of Spinach.ST activation. The fully complementary toehold is denoted as a string of “N” with nucleotide positions numbered above. (C) Detection of RNA CHA circuit output using Spinach.ST. Toehold-mediated strand displacement by a catalyst oligonucleotide leads to the formation of H1:H2 from the kinetically trapped CHA hairpins H1 and H2. The Spinach.ST trigger is appended to H1 such that the toehold domain 2* remains sequestered within the H1 stem, and hence is unable to initiate activation of the Spinach molecular beacon. Upon formation of the H1:H2 circuit output complex, the trigger toehold 2* is exposed and can be quantitated by measuring the fluorescence accumulation of Spinach.ST molecules that undergo conformational activation.