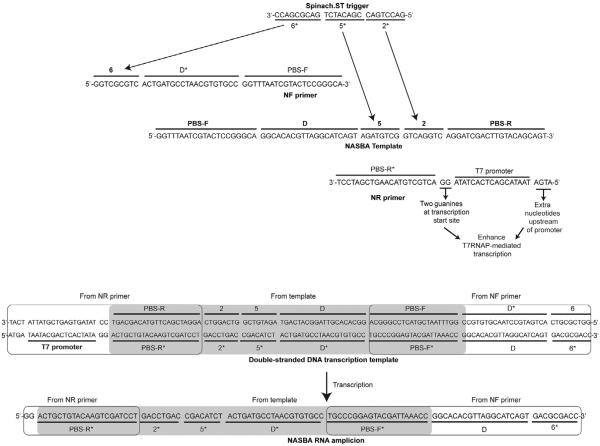
Figure 8.
Examples of nucleic acid sequence-based amplification (NASBA) primer and template design. Sequences complementary to the desired trigger domains 2* and 5* are designated in the templates. Sequences complementary to the trigger domain 6* are appended at the 5′-end of the NF primer. The resulting NASBA RNA amplicons contain the trigger domains 2* and 5* (transcribed from the template) and domain 6* (transcribed from the NF primer). The domain D* is complementary to a stretch of template sequence, termed D, that is inserted in the NF primer. Hybridization of D and D* juxtaposes the otherwise distributed trigger domains via hairpin formation (see Fig. 7). Sequences for domains 2*, 5*, and 6* are derived from the trigger sequence for Spinach.ST2 molecular beacon with the sequence GGA AGC GGT GGC TCA ATG GTA GAG CTT TCG ACG ACA TCT GAC GCG ACC GAA ATG GTG AAG GAC GGG TCC AGT GCT TCG GCA CTG TTG AGT AGA GTG TGA GCT CCG TAA CTG GTC GCG TCG GTC GCG TCA GAT GTC GGT CAG GTC TCG AAG GGT TGC AGG TTC AAT TCC TGT CCG TTT C (Bhadra & Ellington, 2014b). For the synthetic target primer-binding sites (PBS-F and PBS-R) with approximately 50% GC content were used; this was also the case for the hybridization domains (D and its complement D*). The strong 17-mer T7 RNA polymerase promoter sequence (including the two guanine nucleotides at the transcription initiation site) was obtained from literature (Dunn & Studier, 1983; Martin & Coleman, 1987; Milligan et al., 1987). An additional four nucleotide sequence was included at the 5′-end of the promoter based on reports that suggested that T7 polymerase contacts DNA through the −21 position (Baklanov, Golikova, & Malygin, 1996; Ikeda & Richardson, 1986).