Fig. 4.
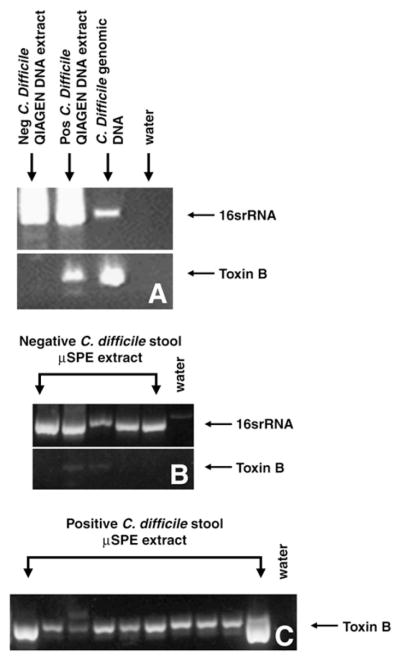
DNA was extracted from C. difficile negative (neg) and positive (pos) stool samples using a Qiagen DNA Stool Mini Kit and PCR was run with both sets of primers (A). All samples were positive for 16srRNA confirming that lysis occurred and that only toxin containing samples were positive for C. difficile toxin B. For extraction using the microfluidic columns (μSPE), no prominent toxin B amplification was observed in negative stool (B) although the microfluidic columns were able to extract DNA (16srRNA). Faint amplification of toxin B was present in 2 of 5 negative stool samples. C. difficile toxin B was detected in DNA extracts from all 9 positive samples using SPE channels (C). Each lane represents separate samples.
