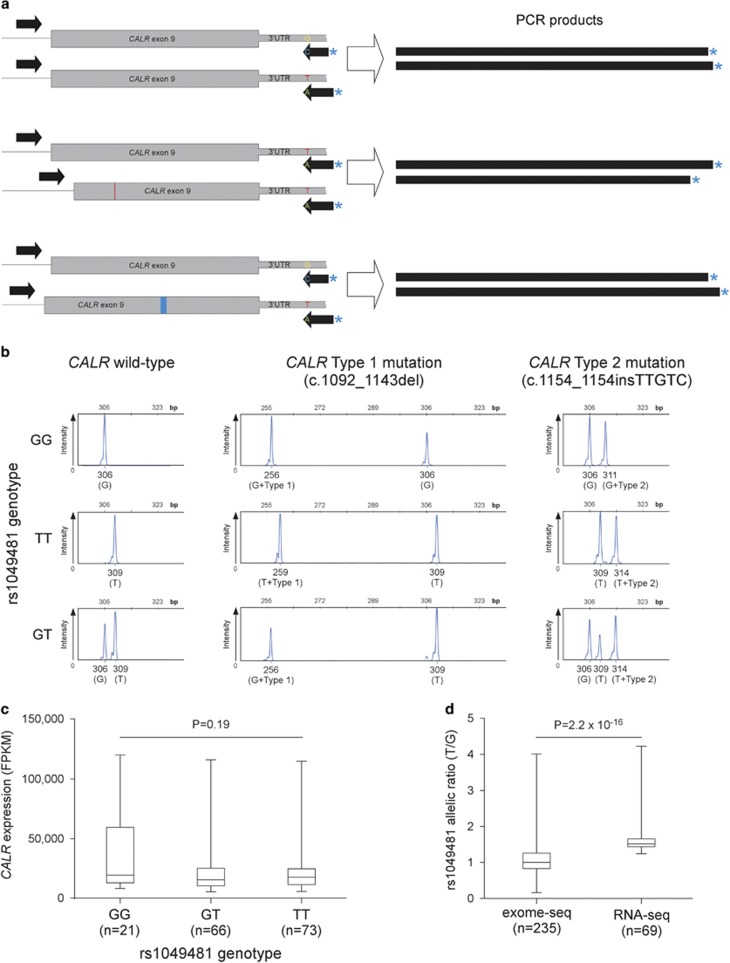
Figure 1.
Investigation of potential allelic imbalances in CALR mutational acquisition and gene expression. (a) Assay used for simultaneous genotyping of rs1049481 and allelic localization of CALR mutations. The two allele-specific labeled reverse primers bind and amplify the chromosomal region bearing the complementary allele of the SNP resulting in products of different sizes. In addition, the size difference of CALR insertions and deletions allows for identification of the allele on which the mutations are acquired. (b) Capillary electrophoresis tracks of the amplified labeled product for different cases of rs1049481 genotypes and CALR mutations. Below each peak, the size, the corresponding genotype (in brackets) and the CALR mutational status are indicated. Coexistence of mutated and wild-type CALR on the T allele suggests incomplete clonality (bottom right panel). (c) The relationship between rs1049481 genotypes and CALR mRNA expression levels. RNA-Seq data on peripheral blood from 173 AML patients were downloaded from The Cancer Genome Atlas (TCGA) project (LAML data set). Normalized expression values were grouped by rs1049481 genotype. There is no detectable statistically significant difference in genotype-specific gene expression (P=0.19; Kruskal–Wallis test). (d) Evidence for allelic expression imbalance at the CALR locus. RNA-Seq and exome sequencing alignment data were downloaded from the TCGA data platform and the 1000 Genomes Project, respectively. Genomic DNA for exome sequencing was derived from normal control tissues, whereas RNA-Seq data was generated from AML peripheral blood enriched for myeloid progenitor cells. Allelic read depth ratios at rs1049481 (T/G) of heterozygous samples that passed genotyping quality filtering are shown. While the T/G ratio of exome sequencing calls is around 1, allelic ratios at RNA level significantly deviate from allelic balance (P=2.2 × 10−16; Mann–Whitney U-test).