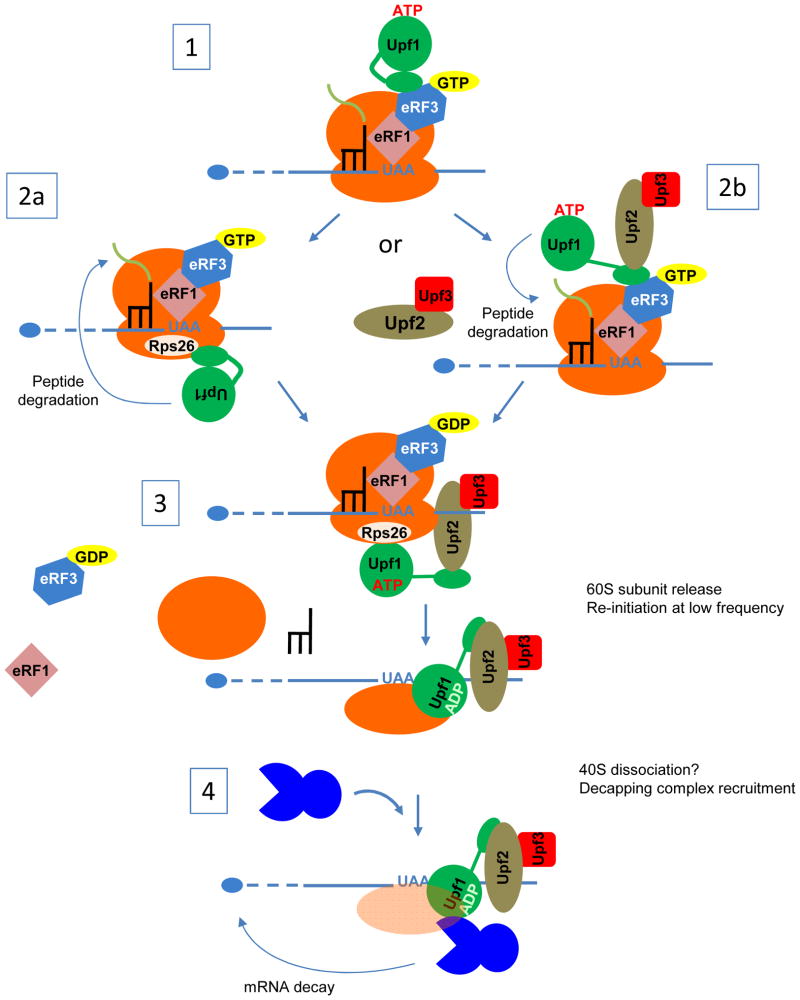
Figure 2. Proposed mechanism for NMD in yeast.
This model is speculative and aims to account for multiple observations on NMD in a uniform scenario. 1. eRF1 and eRF3 are recruited to a ribosome paused at a stop codon in an inappropriate 3′ mRNP context. This leads to improper termination, possibly linked to the inability of eRF3 to leave the complex, and could be favored by the absence of Pab1. Upf1 in an inactive form is recruited to eRF3-eRF1. Two alternative outcomes are then considered: 2a. After its recruitment to the termination complex, Upf1 binds to Rps26 and is joined by Upf2 and Upf3 and “opened” for activation. This step promotes peptide degradation (as indicated by the arrow). 2b. Upf2-Upf3 binds to Upf1 on the termination complex and activates Upf1. The complex then associates with Rps26 through the Upf1 CH domain, and promotes peptide degradation (as indicated by the arrow). 3. Upf1 is active, associated with Upf2-Upf3, and bound to the ribosome via Rsp26. These components promote dissociation of the termination complex, a step that requires the Upf1 helicase activity and leaves the 40S subunit still bound to the mRNA (the events leading to 40S removal are unknown). This step is different from the “normal” recycling event involving Rli1 (see Figure 1). 4. The presence of Upf1 promotes mRNA decapping by Dcp1-Dcp2 (as indicated by the arrow), via Upf1 interaction with Dcp2. Destabilization of the mRNA closed-loop structure, subsequent to improper recycling, could also promote mRNA decay.