Abstract
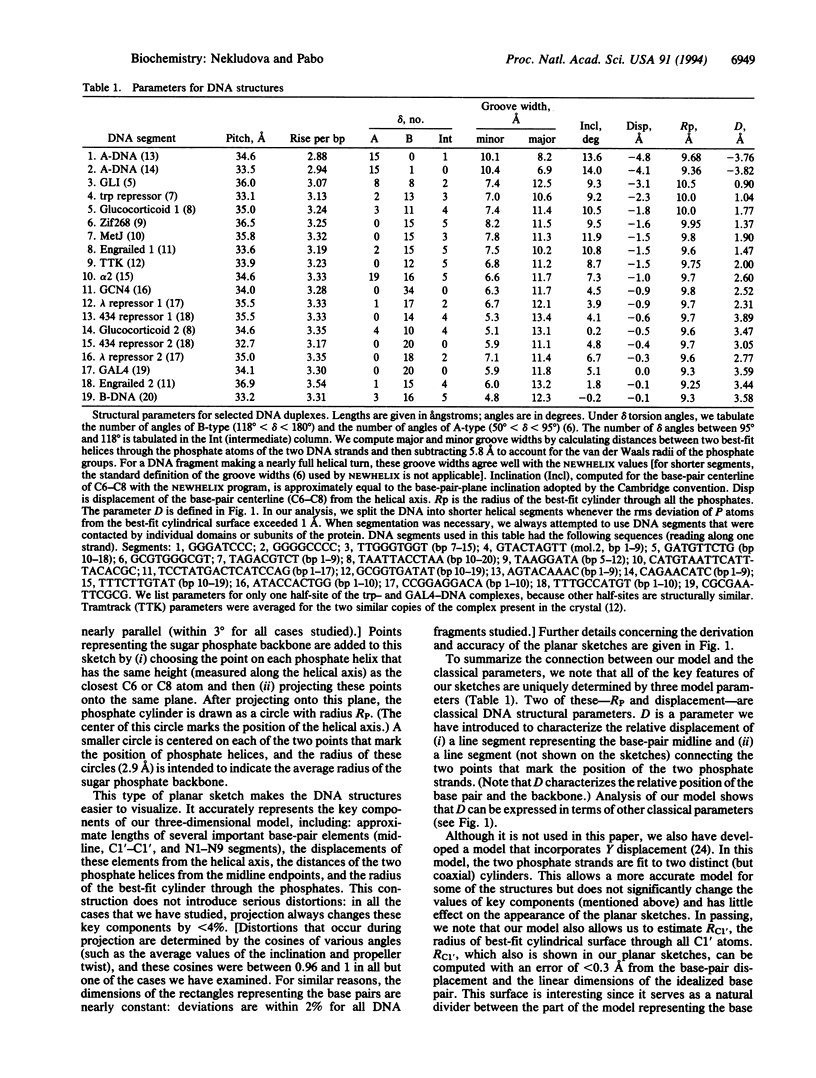
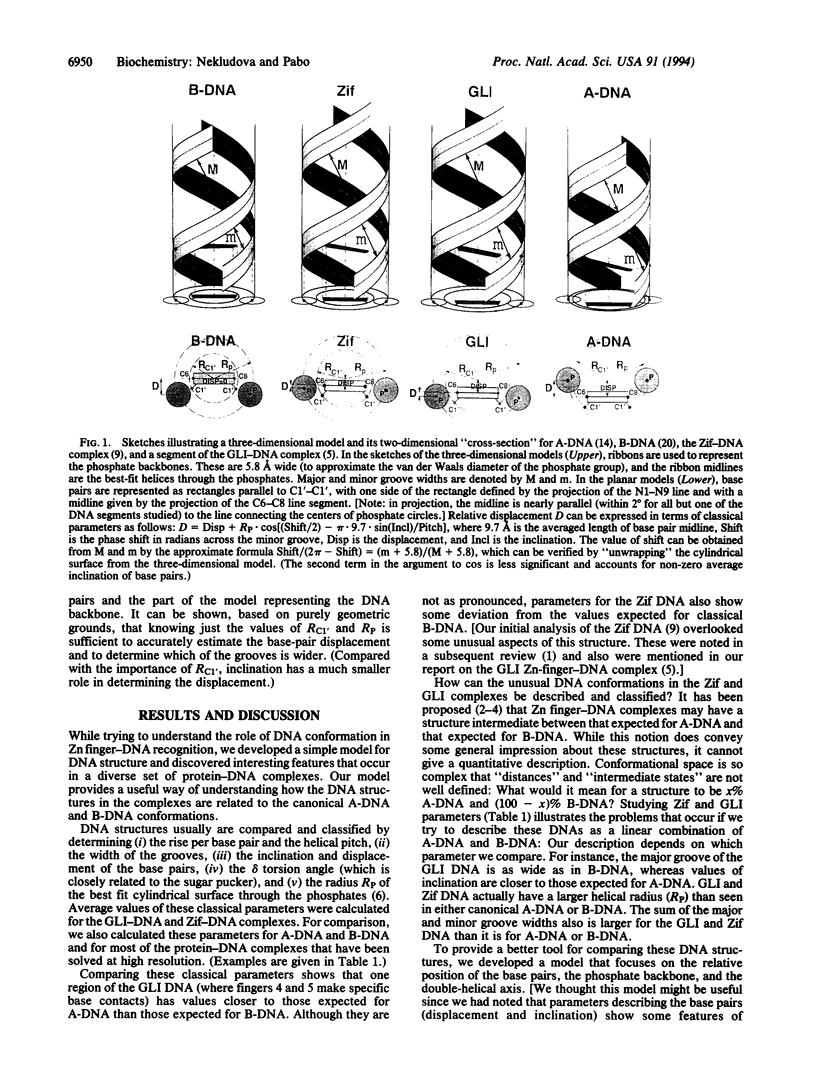
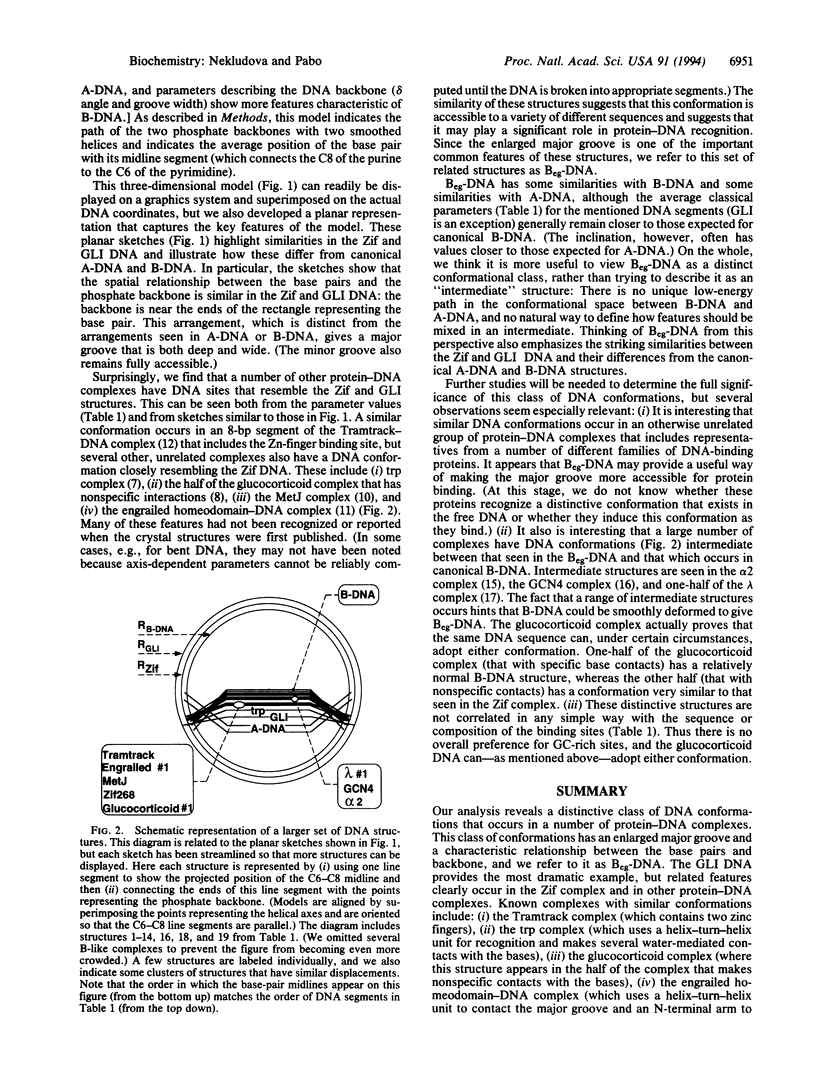
We have analyzed DNA conformations in a series of protein-DNA complexes, and we find that a distinctive conformation--with an enlarged major groove--occurs in a number of different complexes. During this analysis, we also developed a simplified model of DNA structure that illustrates the relative position of (i) the base pairs, (ii) the phosphate backbone, and (iii) the double-helical axis. This model highlights the key structural features of each duplex, facilitating the analysis and comparison of structures that are distinct from canonical A-DNA or B-DNA. Comparing DNA conformations in this way revealed that an otherwise unrelated set of protein-DNA complexes have interesting structural similarities, including an enlarged major groove. We refer to this class of structures as Beg-DNA (where eg means enlarged groove). Since related features occur in such a diverse set of protein-DNA complexes, we suggest that this conformation may have a significant role in protein-DNA recognition.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aggarwal A. K., Rodgers D. W., Drottar M., Ptashne M., Harrison S. C. Recognition of a DNA operator by the repressor of phage 434: a view at high resolution. Science. 1988 Nov 11;242(4880):899–907. doi: 10.1126/science.3187531. [DOI] [PubMed] [Google Scholar]
- Beamer L. J., Pabo C. O. Refined 1.8 A crystal structure of the lambda repressor-operator complex. J Mol Biol. 1992 Sep 5;227(1):177–196. doi: 10.1016/0022-2836(92)90690-l. [DOI] [PubMed] [Google Scholar]
- Definitions and nomenclature of nucleic acid structure parameters. EMBO J. 1989 Jan;8(1):1–4. doi: 10.1002/j.1460-2075.1989.tb03339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew H. R., Wing R. M., Takano T., Broka C., Tanaka S., Itakura K., Dickerson R. E. Structure of a B-DNA dodecamer: conformation and dynamics. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2179–2183. doi: 10.1073/pnas.78.4.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenberger T. E., Brandl C. J., Struhl K., Harrison S. C. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein-DNA complex. Cell. 1992 Dec 24;71(7):1223–1237. doi: 10.1016/s0092-8674(05)80070-4. [DOI] [PubMed] [Google Scholar]
- Fairall L., Martin S., Rhodes D. The DNA binding site of the Xenopus transcription factor IIIA has a non-B-form structure. EMBO J. 1989 Jun;8(6):1809–1817. doi: 10.1002/j.1460-2075.1989.tb03575.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairall L., Schwabe J. W., Chapman L., Finch J. T., Rhodes D. The crystal structure of a two zinc-finger peptide reveals an extension to the rules for zinc-finger/DNA recognition. Nature. 1993 Dec 2;366(6454):483–487. doi: 10.1038/366483a0. [DOI] [PubMed] [Google Scholar]
- Hegde R. S., Grossman S. R., Laimins L. A., Sigler P. B. Crystal structure at 1.7 A of the bovine papillomavirus-1 E2 DNA-binding domain bound to its DNA target. Nature. 1992 Oct 8;359(6395):505–512. doi: 10.1038/359505a0. [DOI] [PubMed] [Google Scholar]
- Huber P. W., Morii T., Mei H. Y., Barton J. K. Structural polymorphism in the major groove of a 5S RNA gene complements the zinc finger domains of transcription factor IIIA. Proc Natl Acad Sci U S A. 1991 Dec 1;88(23):10801–10805. doi: 10.1073/pnas.88.23.10801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissinger C. R., Liu B. S., Martin-Blanco E., Kornberg T. B., Pabo C. O. Crystal structure of an engrailed homeodomain-DNA complex at 2.8 A resolution: a framework for understanding homeodomain-DNA interactions. Cell. 1990 Nov 2;63(3):579–590. doi: 10.1016/0092-8674(90)90453-l. [DOI] [PubMed] [Google Scholar]
- König P., Richmond T. J. The X-ray structure of the GCN4-bZIP bound to ATF/CREB site DNA shows the complex depends on DNA flexibility. J Mol Biol. 1993 Sep 5;233(1):139–154. doi: 10.1006/jmbi.1993.1490. [DOI] [PubMed] [Google Scholar]
- Lauble H., Frank R., Blöcker H., Heinemann U. Three-dimensional structure of d(GGGATCCC) in the crystalline state. Nucleic Acids Res. 1988 Aug 25;16(16):7799–7816. doi: 10.1093/nar/16.16.7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luisi B. F., Xu W. X., Otwinowski Z., Freedman L. P., Yamamoto K. R., Sigler P. B. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature. 1991 Aug 8;352(6335):497–505. doi: 10.1038/352497a0. [DOI] [PubMed] [Google Scholar]
- Marmorstein R., Carey M., Ptashne M., Harrison S. C. DNA recognition by GAL4: structure of a protein-DNA complex. Nature. 1992 Apr 2;356(6368):408–414. doi: 10.1038/356408a0. [DOI] [PubMed] [Google Scholar]
- McCall M., Brown T., Hunter W. N., Kennard O. The crystal structure of d(GGATGGGAG): an essential part of the binding site for transcription factor IIIA. Nature. 1986 Aug 14;322(6080):661–664. doi: 10.1038/322661a0. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Schevitz R. W., Zhang R. G., Lawson C. L., Joachimiak A., Marmorstein R. Q., Luisi B. F., Sigler P. B. Crystal structure of trp repressor/operator complex at atomic resolution. Nature. 1988 Sep 22;335(6188):321–329. doi: 10.1038/335321a0. [DOI] [PubMed] [Google Scholar]
- Pavletich N. P., Pabo C. O. Crystal structure of a five-finger GLI-DNA complex: new perspectives on zinc fingers. Science. 1993 Sep 24;261(5129):1701–1707. doi: 10.1126/science.8378770. [DOI] [PubMed] [Google Scholar]
- Pavletich N. P., Pabo C. O. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991 May 10;252(5007):809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- Schultz S. C., Shields G. C., Steitz T. A. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science. 1991 Aug 30;253(5023):1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- Somers W. S., Phillips S. E. Crystal structure of the met repressor-operator complex at 2.8 A resolution reveals DNA recognition by beta-strands. Nature. 1992 Oct 1;359(6394):387–393. doi: 10.1038/359387a0. [DOI] [PubMed] [Google Scholar]
- Wolberger C., Vershon A. K., Liu B., Johnson A. D., Pabo C. O. Crystal structure of a MAT alpha 2 homeodomain-operator complex suggests a general model for homeodomain-DNA interactions. Cell. 1991 Nov 1;67(3):517–528. doi: 10.1016/0092-8674(91)90526-5. [DOI] [PubMed] [Google Scholar]



