Fig. 4. Viral dynamics during the 2014 outbreak.
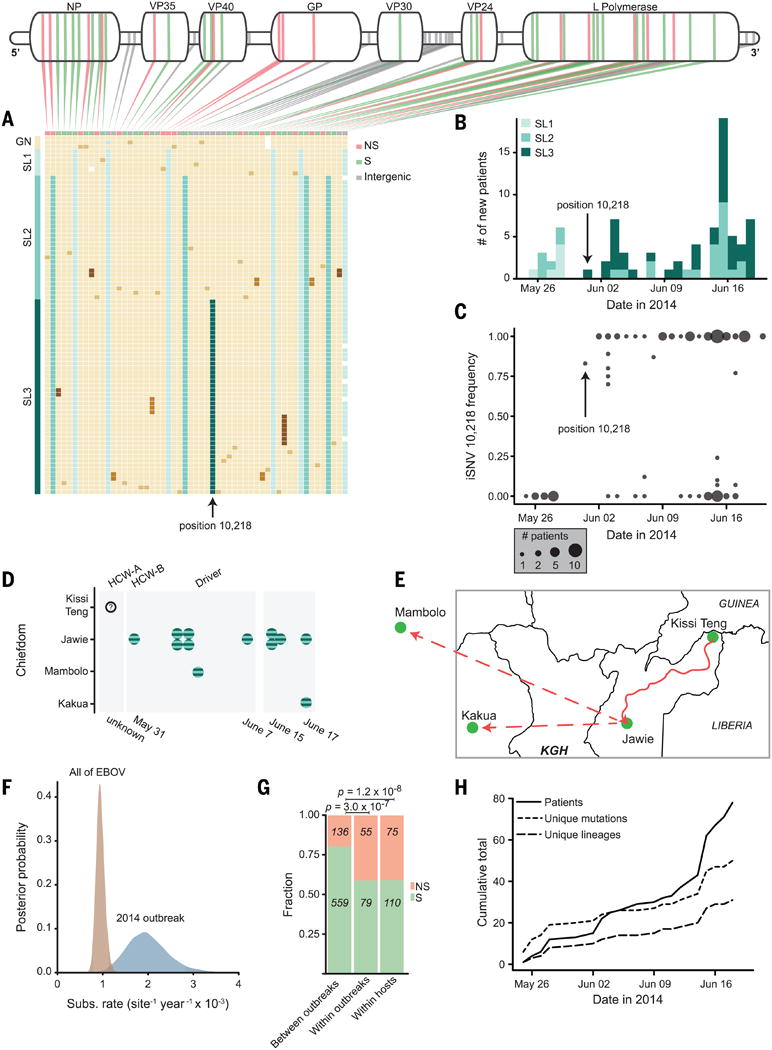
(A) Mutations, one patient sample per row; beige blocks indicate identity with the Kissidougou Guinean sequence (GenBank accession KJ660346).The top row shows the type of mutation (green, synonymous; pink, nonsynonymous; gray, inter-genic), with genomic locations indicated above. Cluster assignments are shown at the left. (B) Number of EVD-confirmed patients per day, colored by cluster. Arrow indicates the first appearance of the derived allele at position 10,218, distinguishing clusters 2 and 3. (C) Intrahost frequency of SNP 10,218 in all 78 patients (absent in 28 patients, polymorphic in 12, fixed in 38). (D and E) Twelve patients carrying iSNV 10,218 cluster geographically and temporally (HCW-A = unse-quenced health care worker; Driver drove HCW-A from Kissi Teng to Jawie, then continued alone to Mambolo; HCW-B treated HCW-A). KGH = location of Kenema Government Hospital. (F) Substitution rates within the 2014 outbreak and between all EVD outbreaks. (G) Proportion of nonsynonymous changes observed on different time scales (green, synonymous; pink, nonsynonymous). (H) Acquisition of genetic variation over time. Fifty mutational events (short dashes) and 29 new viral lineages (long dashes) were observed (intrahost variants not included).
