Figure 1.
NEDD8 and the Neddylation Machinery Accumulate at Sites of DNA Breaks and Promote Cell Survival after NHEJ
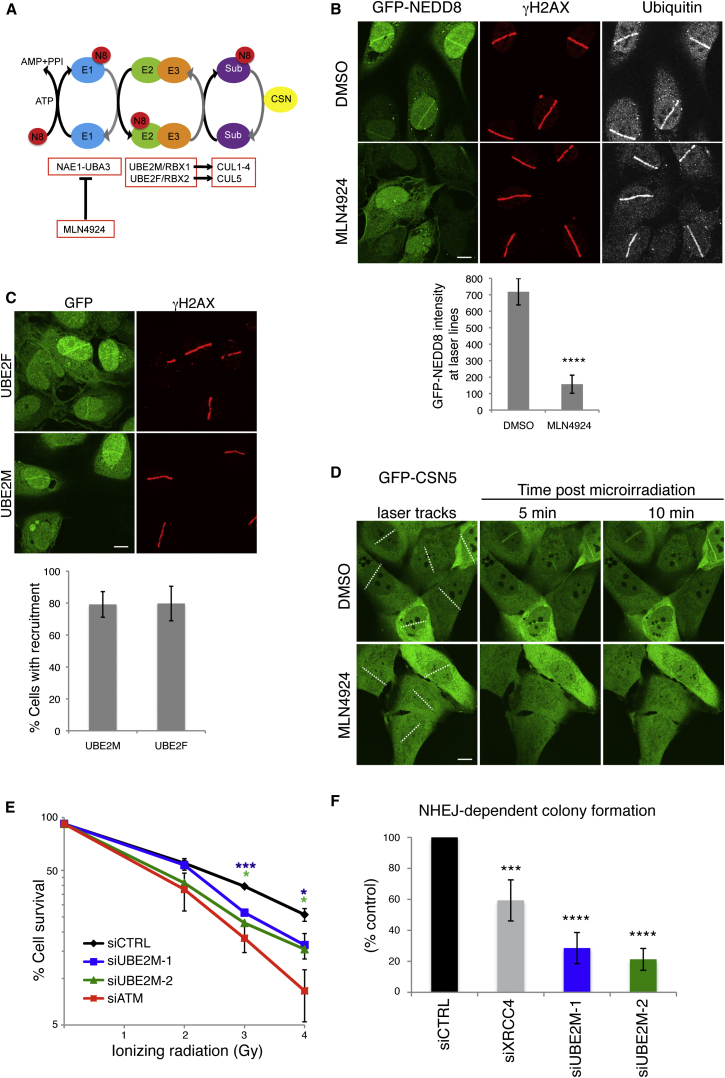
(A) Representation of major neddylation pathway components. NEDD8 (N8) is conjugated in an ATP-dependent cascade involving an E1 (NAE1-UBA3), E2 (UBE2M or F), and E3 (RBX1 or 2) to Cullin substrates (Sub). Neddylation is reversed by the CSN complex. MLN4924 inhibits UBA3. Figure adapted from Brown and Jackson (2015).
(B) MLN4924 blocks NEDD8, but not ubiquitin recruitment to DNA-damage sites. U2OS-GFP-NEDD8 cells were pre-treated for 1 hr with DMSO or 3 μM MLN4924 and laser microirradiated. Cells were fixed after 20 min and visualized by immunofluorescence as indicated. Graph shows average intensity of GFP-NEDD8 at the laser line from three experiments ±SD. White bar represents 10 μM. Asterisks indicate statistically significant difference to control (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p ≤ 0.0001).
(C) GFP-UBE2F and GFP-UBE2M are recruited to DNA-damage sites. U2OS cells stably expressing GFP-UBE2F or GFP-UBE2M were laser microirradiated, fixed, and visualized as in (B). Graph shows average percentage of γH2AX positive cells with detectable GFP-UBE2M or GFP-UBE2F recruitment from five independent experiments ±SD. White bar represents 10 μM.
(D) GFP-CSN5 recruitment to DNA-damage sites is blocked by MLN4924. U2OS cells stably expressing GFP-CSN5 were treated as in (B). Images were acquired by live cell imaging. Laser tracks are indicated by dashed white lines. White bar represents 10 μM
(E) UBE2M depletion causes hypersensitivity to IR. Clonogenic U2OS cell survivals were performed after transfection with indicated siRNAs and doses of IR. Each point represents an average of at least three independent experiments (except UBE2M-2 which was repeated twice). Error bars correspond to SDs, and asterisks are as in (B).
(F) UBE2M depletion causes an NHEJ defect. Random plasmid integration assay was performed in U2OS cells transfected with indicated siRNAs. Error bars correspond to SD of at least three independent experiments (asterisks as in B). See also Figure S1.