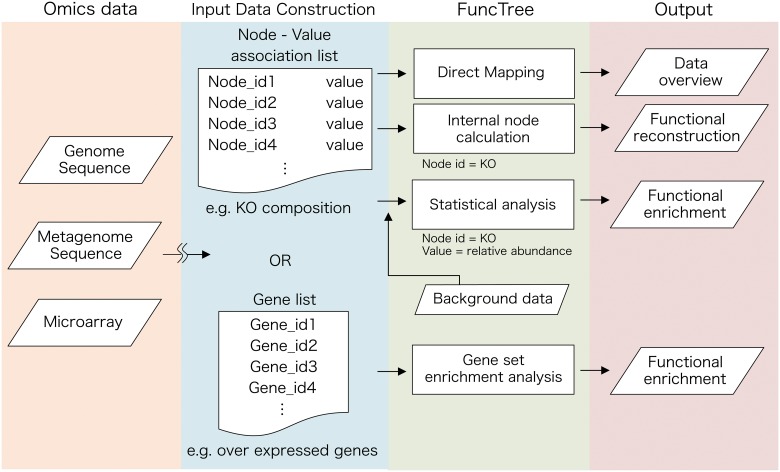
Fig 2. Flowchart for data mapping using FuncTree.
FuncTree is a web-based application that is capable of analyzing and visualizing the functional characteristic of large-scale omics data. As an input, FuncTree expects either a list of node-value association, which associates each node with a designated value, or a list of genes. Value for each node would have to be calculated locally by user, and may represent a number of different variables such as relative abundance. Gene list may represent a list of over-expressed genes in a microarray experiment. Depending on the Input, FuncTree is able to perform various visualizations such as, (i) Direct mapping: directly map the value onto the designated node, (ii) Internal node calculation: Calculate the functional potential of the upper four layer from KO composition, (iii) Statistical analysis: Conduct enrichment analysis for each layer from KO composition against a predefined background data of several human metagenomic dataset, and (iv) Gene set enrichment analysis: Conduct enrichment analysis for each layer from gene list.