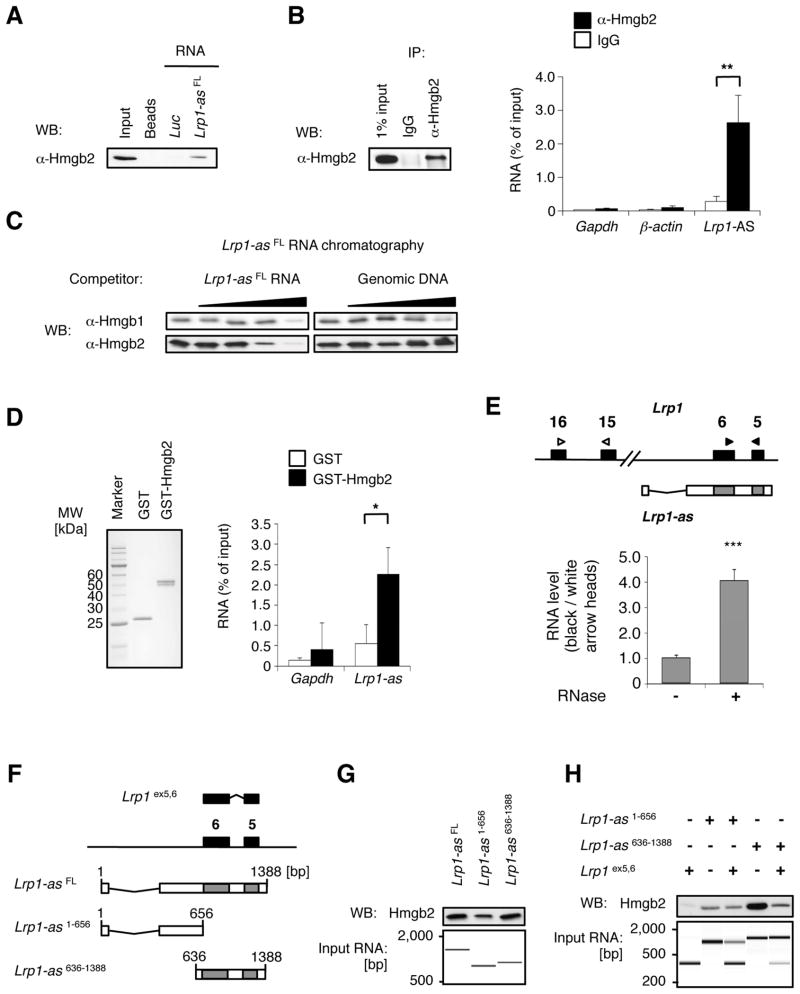
Figure 3. Lrp1-AS binds to Hmgb2.
(A) RNase-assisted RNA chromatography on full-length Lrp1-AS FL in RAW264.7 nuclear extracts, visualized by Western blotting (WB) with specific antibody against Hmgb2.
(B) Immunoprecipitation with control IgG or specific antibody against Hmgb2 from RAW264.7 lysates, visualized by WB (left), and co-precipitated RNAs from (B) were detected by qRT-PCR using primer pairs for Lrp1-AS, Gapdh or β-actin (right).
(C) In vitro pulldown of 6XHis-tagged Hmgb2 with Lrp1-ASFL and free nucleic acids as indicated.
(D) In vitro pulldown of endogenous Lrp1-AS with GST or GST-fused Hmgb2 from RAW264.7 nuclear extracts. RNAs were detected by qRT-PCR using primer pairs for Lrp1-AS or Gapdh (right). Equal input proteins were visualized on electrophoresis by CBB stain (left). Mean ± s.d. (n = 3 replicates) are shown in all bar graphs. *P < 0.05, **P < 0.01 determined by ANOVA.
(E) RNase protection assay. The RNA ratios were calculated from qRT-PCR using primer pairs for overlap region (black arrowhead) or non overlap region (white arrowhead) from RAW264.7 nuclear extracts treated with (+) or without (−) RNase A. Mean ± s.d. (n = 3 replicates) are shown. ***P < 0.001 determined by ANOVA.
(F) Schematic illustration of deletion constructs of Lrp1 and Lrp1-AS, used for (C) and (D): Lrp1 construct contains exons 5 and 6 (395 bp) of Lrp1. Lrp1-AS636-1388 construct contains the overlap region between Lrp1 and Lrp1-AS, whereas Lrp1-AS1-656 only contains the 5′ fragment of Lrp1-AS.
(G) RNA chromatography on full length or fragments of Lrp1-AS, as indicated above, in RAW264.7 nuclear extracts, followed by Western blotting (WB, upper). Equal input RNAs were visualized on electrophoresis by Agilent Bioanalyzer (lower).
(H) RNA chromatography on Lrp1-AS fragments, Lrp1 fragment, or Lrp1-AS:Lrp1 fragments hybrid, as indicated above, in RAW264.7 nuclear extracts, followed by Western blotting (WB, upper). Equal input RNAs were visualized on electrophoresis by Agilent Bioanalyzer (lower).