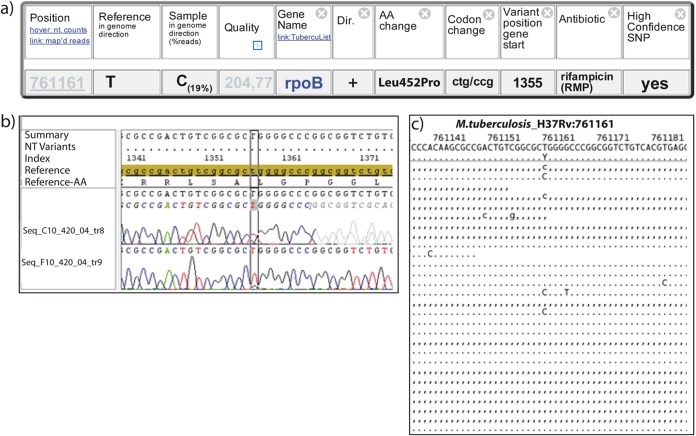
FIG 4.
Detection of heteroresistant samples with PhyResSE. (a) Developing RIF resistance. Only 19% of the reads show CCG (Pro) instead of the wild-type codon CTG (Leu). This reflects the small proportion of bacteria already carrying RIF resistance at the time of sample preparation. Dir., direction; AA, amino acid. (b) Electropherogram of Sanger sequencing at position 761161. In the reverse sequence of 420/04 (Tr8), no additional peak is visible. In the forward sequence (Tr9), a small C peak is visible. NT, nucleotide. (c) Mapping result at position 761161. Approximately 19% of the reads show a C instead of the wild-type base T (independent of the strand). A representative subset of 26 out of 102 reads in total is shown for clarity.