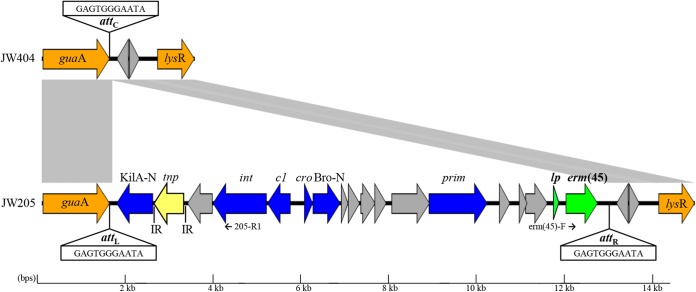
FIG 1.
Schematic gene map showing the erm(45)-containing island and flanking region of S. fleurettii JW205 (ENA accession number LN680996), as well as the chromosomal glutamine amidotransferase gene (guaA) region of erm(45)-negative strain JW404. Gray areas represent regions showing high similarity at nucleotide level (>98%). Arrows represent position and orientation of open reading frames (ORFs). The erm(45) and its leader peptide (lp) gene are shown in green. The 11-bp (GAGTGGGAATA) chromosomal hot spot situated within the 3′ end of guaA is abbreviated as attC. This attachment site is illustrated as attL and attR at each extremity of the island. The IS431 transposase gene (tnp) illustrated in yellow is flanked by 16-bp inverted repeats (IR) (GGTTCTGTTGCAAAGT). Other putative genes and functions: KilA-N, KilA-N domain-carrying putative DNA-binding protein; c1/cro, repressors; Bro-N, Bro-N domain-carrying putative DNA-binding protein; lysR, transcriptional regulator; int, integrase; prim, primase. Color code: S. fleurettii core genome, orange; erm(45)-carrying island genes related to SaPI and bacteriophage genomes, blue; antibiotic resistance, green; no known function, significant similarity to proteins in the database used, or any known protein signatures, gray. Primers used to detect circular forms of the erm(45)-carrying island are indicated by small black arrows. The figure was generated using the program Easyfig (25).