FIG 2.
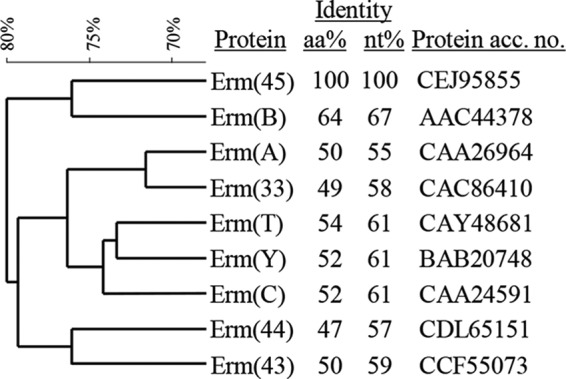
Relatedness of the novel erythromycin resistance methylase Erm(45) with other methylases commonly detected in Staphylococcus species (9, 12). Amino acid (aa) and nucleotide (nt) identity percentages were obtained by Clustal Omega sequence alignment (http://www.ebi.ac.uk/Tools/msa/clustalo/). Erm aa sequence clustering was performed by BioNumerics 7.1 (Applied Maths). Comparison settings used the standard algorithm for pairwise alignment, open gap penalty 100%, unit gap penalty 0%, and the unweighted-pair group method using average linkages.
