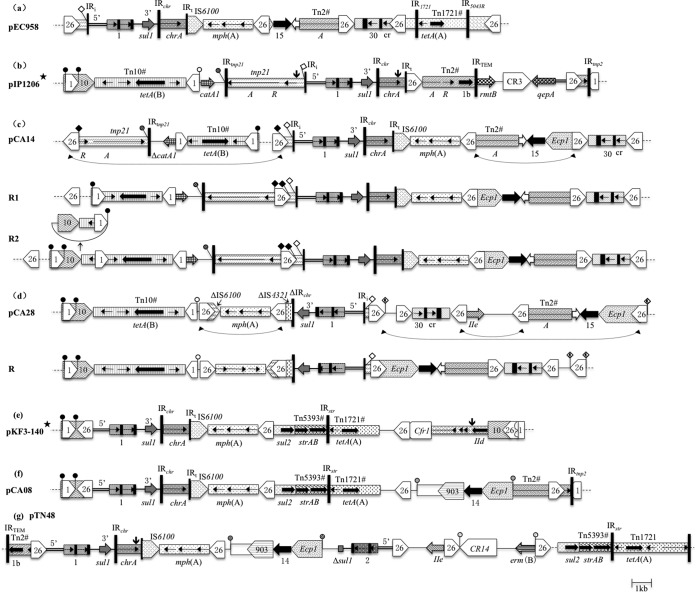
FIG 2.
MRRs of plasmids pCA14 (ST131, blaCTX-M-15), pCA28 (ST44, blaCTX-M-15), and pCA08 (ST131, blaCTX-M-14), compared with those of related IncF plasmids pIP1206 (AM886293) and pKF3-140 (FJ876827), the IncF blaCTX-M-15-harboring plasmid pEC958 (HG941719), and the IncF blaCTX-M-14-harboring plasmid pETN48 (FQ482074). Various transposons and other modules have different shading and are generally labeled once for each plasmid. # indicates that a transposon is incomplete. ISs are pointed boxes labeled with their number/name. Tall bars represent the 38-bp IR of transposons, as indicated. Positions/orientations of selected resistance and other genes are indicated by arrows, generally labeled once for each plasmid. Abbreviations: A, tnpA; R, tnpR; 30, blaOXA-30; cr, aac(6′)-Ib-cr; IIe, aac(3)-IIe; 15, blaCTX-M-15; 1b, blaTEM-1b; 14, blaCTX-M-14; IId, aac(3)-IId. Class 1 integron components are indicated as follows: 5′, 5′-CS; 3′, 3′-CS; small black boxes, attC sites; narrow box 1, dfrA17-aadA5 cassette array; narrow box 2, aacA4-cmlA1 cassette array; IRi, 25-bp IR at intI1 end; IRt, 25-bp IR at tni end. The chrA-mph(A) module is usually located after the 3′-CS, which consists of part of a chromate resistance transposon (IRchr-chrA), the 123-bp IRt end of tni402, IS6100, and the mph(A)-mrx-mphR(A) macrolide resistance gene cluster (9). Paired filled circles represent direct repeats (DRs), and paired squares represent flanking sequences that are reverse complementary to one another. An unpaired circle or square indicates the same boundary between regions present in another of the structures shown. Asterisks against plasmid names indicate that the structures shown have been rearranged to emphasize relationship, usually by inverting regions flanked by IS26 (21). Vertical arrows indicate the positions of IS26 elements with DRs that have been removed for ease of comparison. Dashed lines represent the IncFII backbone. Structures named R1, R2 below CA14, and R below CA28 are hypothetical and are intended to facilitate comparison between related MRRs. Dashed arrowed lines indicate where homologous recombination (HR) happens to invert or delete the regions between two IS26 elements. (c) pCA14 MRR. R1, two inversion events by homologous recombination between IS26 elements in opposite orientations may have occurred (shown by the dashed arrowed lines) in hypothetical structure R1 to yield pCA14 MRR; R2, the circular molecule above R2 may have been deleted in R2 by recombination between IS1 leading to the generation of R1. (d) pCA28 MRR. R, two inversion events by HR between IS26 elements in opposite orientations and a deletion event by recombination between IS26 in the same orientation (shown by the dashed arrowed lines) may have occurred and resulted in hypothetical structure R.