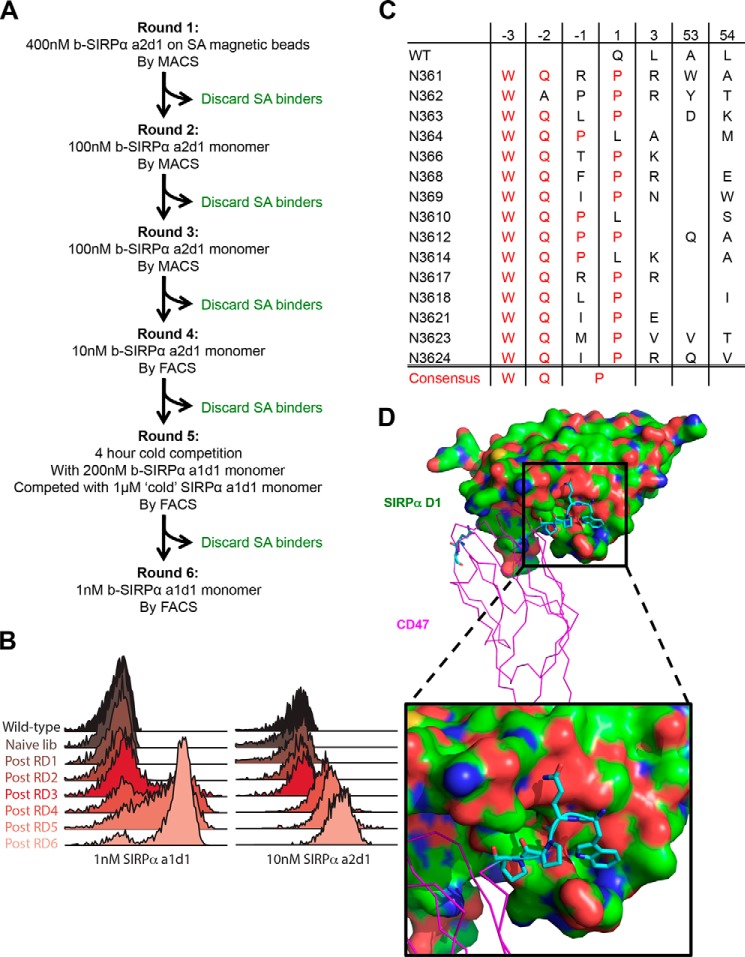
FIGURE 4.
Affinity evolution of CD47 extension library. A, schematics of the selection criteria at each round of the extension library selections. MACS, magnetic activated cell sorting; FACS, fluorescence-activated cell sorting; and SA, streptavidin. B, histogram overlays assessing SIRPα a1d1 (left) and a2d1 (right) staining of the library at each round. C, summary of sequences of engineered CD47 variants. The position of mutated residues and their corresponding sequence in WT is denoted at the top of the table. Positions −3, −2, and −1 denote the “Velcro” residues added to the N terminus. Red text indicates the consensus mutations. D, model of SIRPα-N3612 complex. The structure was modeled using the WT complex structure with PyMOL and Coot and then energy-minimized for 1000 cycles using crystallography and NMR system. SIRPα (green) is shown in surface representation. CD47 (magenta) is shown in ribbon representation, and the mutations (cyan) present in N3612, including the “Velcro”, are shown in stick representation.