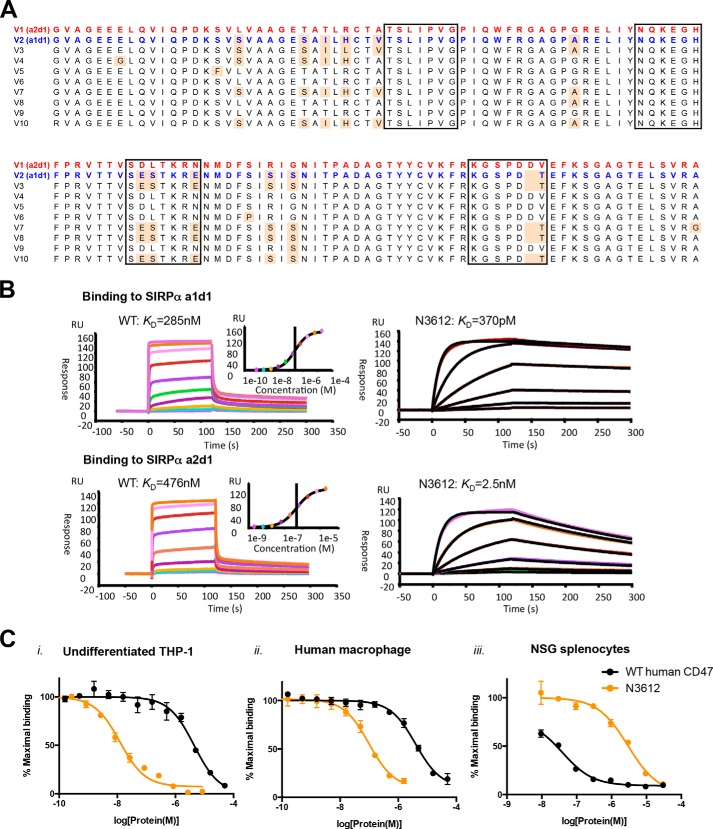
FIGURE 5.
Biophysical characterization of Velcro-CD47. A, amino acid sequence alignment of known human SIRPα-binding domain alleles, showing that there are only two variations, a1d1 or a2d1, at the CD47-contact interface. Red text marks the amino acid sequence of V1 (a2d1), the most prominent human SIRPα allele, and blue text marks that of V2 (a1d1), the second most prominent human SIRPα allele. The black boxes indicate the residues that interact with CD47, and the pale orange shade indicates residues that are different from the V1 sequence. B, representative surface plasmon resonance sensorgrams of WT CD47 and high affinity variant N3612 binding to immobilized SIRPα a1d1 (upper panel) and a2d1 (lower panel). RU, response unit. C, dose-response curves of SIRPα antagonism on panel i, undifferentiated THP-1; panel ii, primary human macrophages; and panel iii, NSG splenocytes with WT human CD47 (blue) and high affinity CD47 variant N3612 (red). Cells were stained with titrating concentrations of SIRPα blocking agents in competition with 100 nm Alexa Fluor 647-conjugated WT human CD47 tetramer for THP-1 cells and human macrophages or WT mouse CD47 tetramer for NSG splenocytes. Data represent mean fluorescence intensity normalized to maximal binding for each cell type ± S.E. of triplicate wells. Data are representative of two independent experiments.