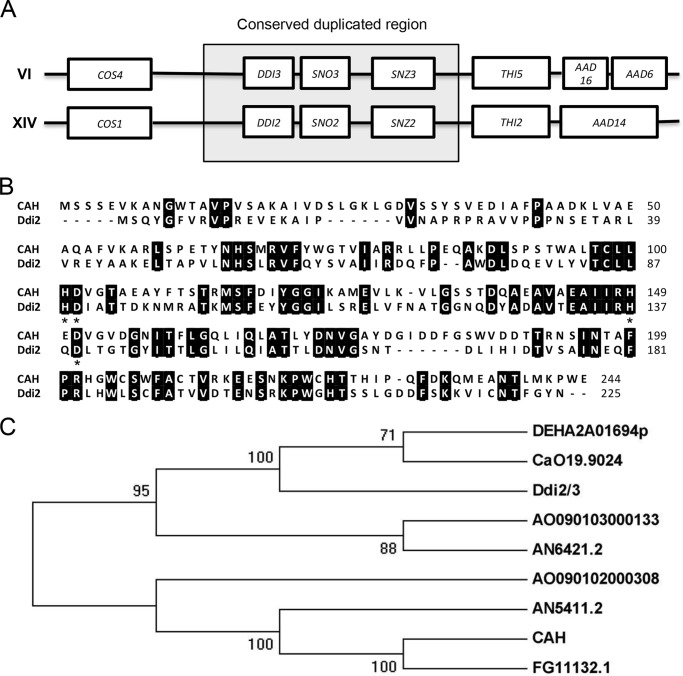
FIGURE 2.
Genomic and protein sequence analyses of Ddi2/3. A, DDI2 and DDI3 genes are located within a highly conserved duplicated region. DDI2 and DDI3 and their flanking 20-kb regions are thought to derive from gene duplication, in which the boxed regions are highly conserved in DNA sequence. B, amino acid sequence alignment of Ddi2/3 with CAH from M. verrucaria. Identical amino acid residues are highlighted. Signature conserved HD residues are marked with asterisks. Protein sequences were retrieved from the NCBI protein database (CAH: AAA33429.1, and Ddi2: P0CH63.1), and the alignment was done with ClustalW. C, phylogenetic tree illustrating the homology among available cyanamide hydratase domain-containing proteins: DEHA2A01694p (Debaryomyces hansenii CBS767), CaO19.9024 (Candida albicans SC5314), AO090103000133 (Aspergillus oryzae RIB40), AN6421.2 (A. nidulans FGSC A4), AO090102000308 (A. oryzae RIB40), AN5411.2 (Aspergillus nidulans FGSC A4), CAH (M. verrucaria) and FG11132.1 (Fusarium graminearum PH-1). Amino acid sequences of proteins were retrieved from the NCBI conserved domain database. MEGA5 was used to predict the phylogenetic tree based on maximum likelihood. The phylogeny was tested by using a bootstrap method, and the bootstrap replications were set at 1000.