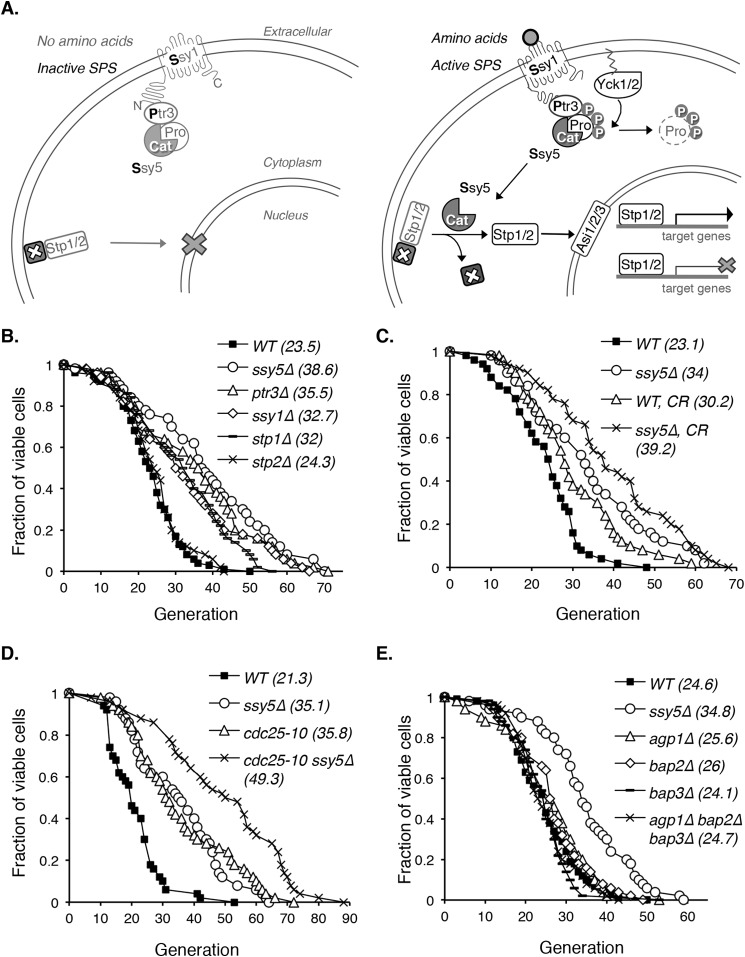
FIGURE 1.
Yeast mutants with reduced SPS signaling show increased replicative life span. A, a simple model of the SPS nutrient-sensing pathway in S. cerevisiae. The left panel shows inactive SPS without amino acid stimulation. Upon activation by extracellular amino acids, SPS activates transcription factors Stp1 and Stp2 (right panel) to modulate (activate or inhibit) downstream gene expression. Pro, Ssy5 prodomain; Cat, Ssy5 catalytic domain; P, phosphorylation. B, deletions of various SPS pathway components extend RLS. The RLS of all mutants (except for stp2Δ) are significantly increased (p < 0.005) when compared with the WT. Δ: gene deletions. C, moderate CR further extends RLS in the SPS mutant ssy5Δ background (ssy5Δ versus ssy5Δ, CR; p < 0.05). CR, 0.5% glucose (versus 2% glucose in standard growth media). D, cdc25-10, a CR genetic mimic, further extends life span in the ssy5Δ background (ssy5Δ versus cdc25-10 ssy5Δ; p < 0.005). E, reduced amino acid uptake is not the main cause of RLS extension in ssy5Δ cells. Unlike ssy5Δ, deletion of SPS downstream amino acid permeases AGP1, BAP2, and BAP3 does not extend RLS. Data shown are representative of multiple independent experiments. Statistical analysis of RLS is determined by the Wilcoxon rank sum test.