FIGURE 1.
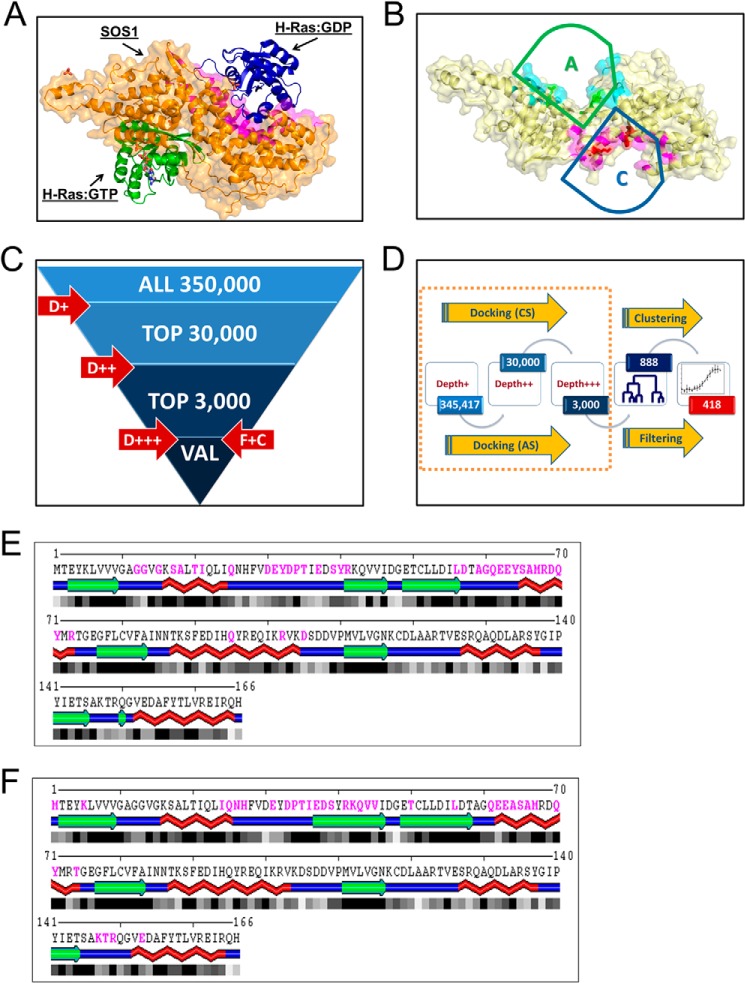
Ensemble docking based virtual screening for candidate small molecule inhibitors of SOS1. A, crystal structure of the REM and Cdc25 catalytic domains of SOS1 in complex with or without both inactive and active H-Ras at the catalytic and allosteric sites of SOS1, respectively (PDB code 1XD2) (19). SOS1 is depicted in orange; inactive H-Ras at the catalytic site is depicted in blue, and active H-Ras at the allosteric site is depicted in green. B, catalytic targeted pocket in SOS1 is highlighted in magenta and indicated by a blue arc-topped triangle labeled with C. The allosteric targeted pocket in SOS1 is highlighted in turquoise and indicated by a green arc-topped triangle labeled with A. Residues at structurally equivalent positions in both pockets are further highlighted by their side chains. C, multistage virtual screening approach is as follows: initial docking performed with limited sampling (D+ or depth 1) to identify top 30,000 compounds, followed by depth 2 (D++) docking with increased sampling to identify top 3000, followed up by docking with extensive sampling (D+++), filtering using predicted inhibition constants and entropy of docking, and clustering based on chemical similarity to identify a set of about 300 compounds in each run. D, overall flow of virtual screening, starting from multiple runs of docking simulations as shown in C and targeting catalytic (CS) or allosteric (AS) sites, using bound or unbound structures of SOS1, followed by selection (filtering) of compounds with best median predicted inhibition constants and low entropy of clustering poses, and by clustering of chemically structurally similar candidates to select a final set of 418 candidates for experimental screening. E and F, SOS1 interaction interface on Ras for both the catalytic and allosteric sites. Ras residues buried upon complex formation with both the SOS1 catalytic and allosteric sites are shown in magenta, as identified by Sppider using default settings. SOS1 interaction interface on Ras for the catalytic site (PDB code 1XD2 and chain B) (E) and SOS1 interaction interface on Ras for the allosteric site (PDB code 1XD2 and chain A) (F). The large overlapping interacting regions on Ras for both the catalytic and allosteric sites, in particular within the switch I region, should be noted.