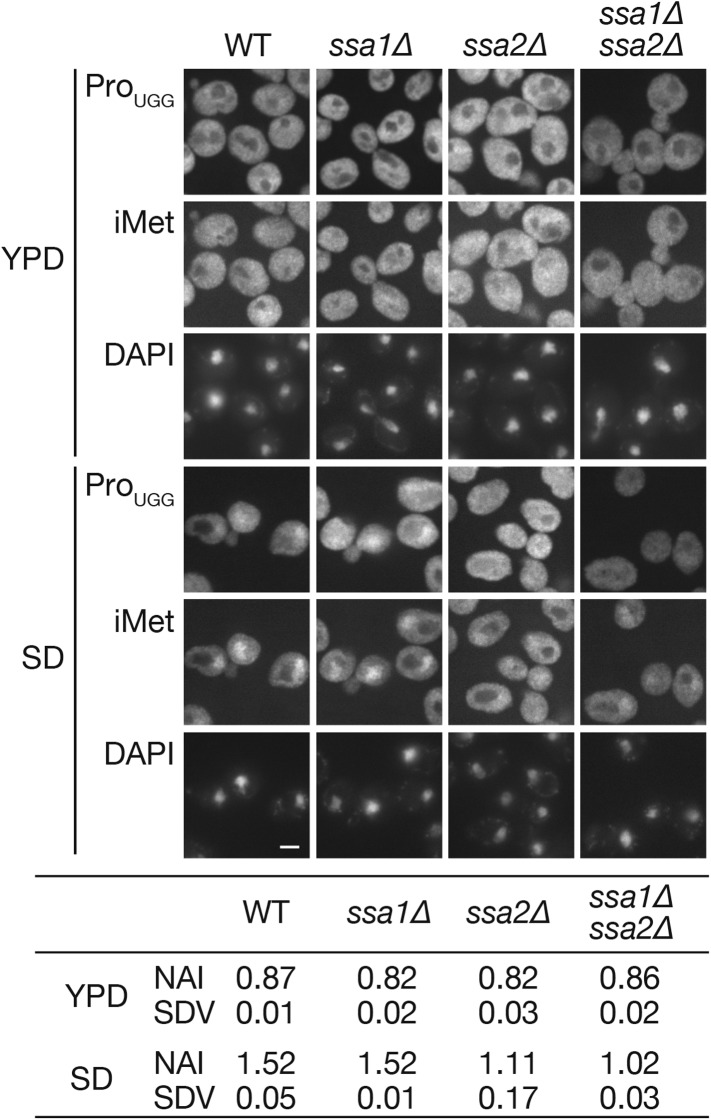
Figure 3. The ssa1∆ ssa2∆ double deletion does not cause a synergistic effect on the nuclear accumulation of tRNAs under starvation conditions.
Wild-type (W303-1A), ssa1∆ (TYSC918), ssa2∆ (TYSC920), and ssa1∆ ssa2∆ double mutant (TYSC1013) strains were cultured in YPD (YPD) and transferred to SD+Ade, Ura (SD) for 2 hr. The cells were subsequently subjected to FISH analysis with anti-tRNA-ProUGG and tRNA-iMet probes. Bar, 5 µm. The fluorescence signals of tRNA-ProUGG images of three independent experiments were quantified, and the average NAIs with SDVs were calculated. Original microscopic images and individual data for quantitative FISH in this figure will be found in Figure 3—source data 1.
DOI: http://dx.doi.org/10.7554/eLife.04659.010