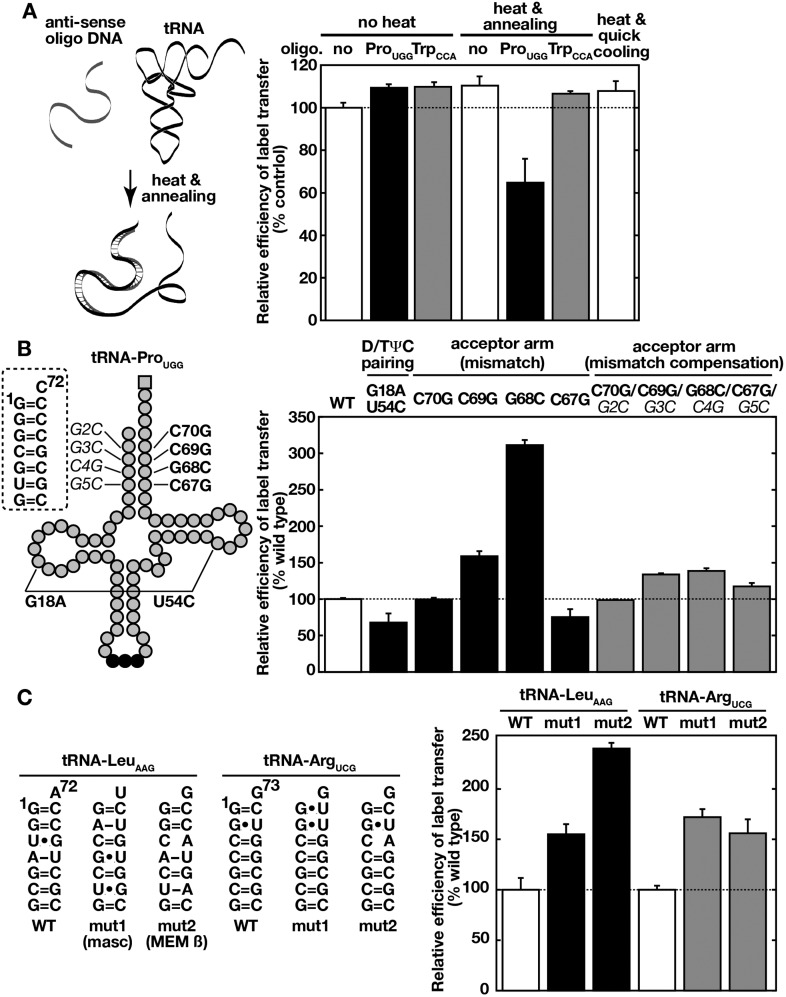
Figure 5. Ssa proteins recognize higher order structures of tRNAs and prefer a destabilized acceptor stem.
(A) 32P-labeled tRNA-ProUGG was mixed without (no, white bar) or, with its anti-sense (ProUGG, black bar) or unrelated control (anti-sense of TrpCCA, gray bar) oligo-DNA. tRNA/oligo-DNA mixtures were heat-denatured and annealed to form DNA-RNA hybrids (‘heat and annealing’) as indicated on the left. A sample without oligo-DNA was also heated and quick-cooled (the far right bar). The resulting samples were then subjected to the label transfer assay with Ssa2p. (B) Label transfer from tRNA-ProUGG mutants was assayed with Ssa2p. Mutation sites of tRNA-ProUGG used in the assay are indicated schematically in the left. Boldface characters indicate mutations introduced to cause destabilization, and italicized characters do mutations that compensate for mismatches caused by the former mutations. The label transfer efficiencies of mutant tRNAs to Ssa2p were summarized in the right graph. (C) Wild-type (WT) and mutant forms (mut1 and mut2) of human tRNA-LeuAAG and tRNA-ArgUCG were subjected to the label transfer assay with Ssa2p. Sequences of the acceptor stems of the tRNAs are shown in the left. The tRNA-LeuAAG derivatives received replacements of the acceptor stem of tRNA-LeuAAG with those of tRNA-like ncRNAs such as MALAT1-associated small cytoplasmic RNA (mascRNA, mut1) and MEM β RNA (mut2). For panels B and C, the label transfer efficiency of the wild-type tRNA was set to 100%. Similar results were obtained with Ssa1p (not shown). Original gel images and individual quantification data for the label transfer assays in this figure will be found in Figure 5—source data 1.
DOI: http://dx.doi.org/10.7554/eLife.04659.015