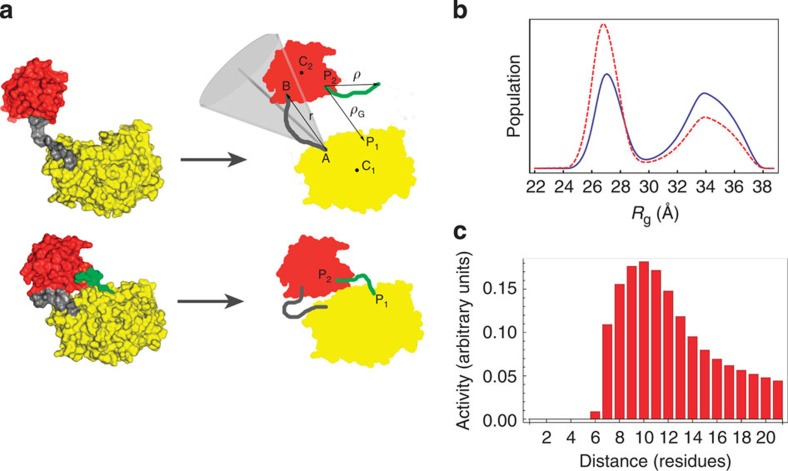
Figure 5. Coarse-grained theoretical model of GalNAc-T2.
(a) The crystal structure of GalNAc-T2-UDP-MUC5AC-13 (bottom left) is taken as a reference for the model (bottom, right). The apo form, without the peptide (green), is obtained by removing the latter from the coordinates file. Upon fixing the relative orientation of the two domains (red and yellow for the lectin and catalytic domains, respectively), protein conformations depend on the flexible linker (grey), modelled as a (non-extendable) semi-flexible chain with an attractive interaction between its ends. All other conformations of the protein (top left) are modelled on the basis of the previous one, by letting the end-to-end vector (r) explore the region within a cone (light grey) of angular amplitude 2θ0 (see Supplementary Methods for details). In the holo-form, the peptide (green) is modelled as a WLC of length (l) residues (the distance between the glycosylating sites on the ligand), bound at P2 on the lectin domain. The enzymatic reaction can take place in any protein conformation providing that the ligand free-end finds correct place P1 on the catalytic domain. (b) Model predictions for the probability distribution [G(Rg)] of the radius of gyration Rg. The apo-form is shown as a blue solid line, whereas the holo-form complexed with peptide is shown as a red dashed line. (c) Enzymatic activity [σ(l,lc)] as a function of the residue separation (l) between potential glycosites and a prior fixed glycosite of the glycopeptides (see Supplementary Methods).