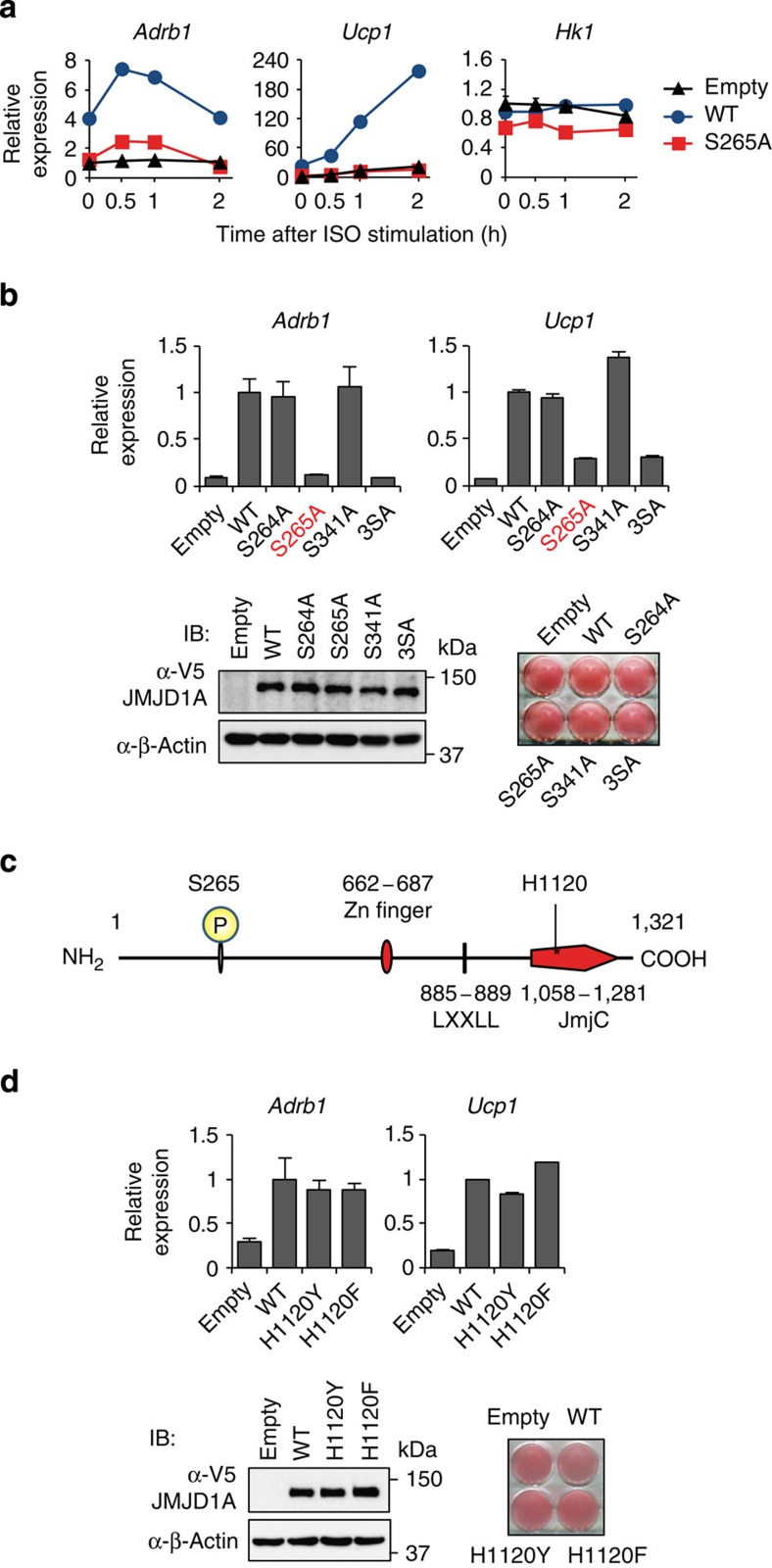
Figure 3. Phosphorylation of JMJD1A at S265 is crucial for β-adrenergic-induced gene transcriptions.
(a) ISO-induced Adrb1 and Ucp1 mRNA levels in iBATshs stably expressing WT- or S265A-hJMJD1A or empty vector were measured by RT-qPCR. The mRNA values are depicted relative to mRNA in iBATshs transduced with empty vector on day 8 of differentiation before ISO treatment (0 h), which are arbitrarily defined as 1. (b) Adrb1 and Ucp1 mRNA levels in WT or serine to alanine mutants (S264A, S265A, S341A or, 3SA) JMJD1A expressing iBATshs measured by RT-qPCR after 1 h ISO treatment (top panel). 3SA represents all three mutations of S264A, S265A and S341A. Data were presented as fold change relative to WT-hJMJD1A-iBATshs after normalized to cyclophilin. Immunoblot (IB) analysis for WT and various mutant JMJD1A proteins and Oil Red O (ORO) staining (bottom panel). (c) Schematic representation of the domain architecture of hJMJD1A. Phosphorylation site at S265 and Fe(II) binding site at H1120 are shown. (d) Comparable ISO-induced gene expressions of Adrb1 and Ucp1 in WT and demethylase dead JMJD1A mutants expressing iBATshs. RT-qPCR was performed to quantify mRNA levels of Adrb1 and Ucp1 genes in WT-, H1120Y- or H1120F-hJMJD1A- iBATshs treated with ISO for 1 h (top panel). Data were presented as fold change relative to WT-hJMJD1A-iBATshs. IB analysis and ORO in the indicated iBATs (bottom panel). Data are presented as mean±s.e.m. of three technical replicates (a,b,d) (error bars are too tiny to see in some figures). Uncropped images of the blots (b,d) are shown in Supplementary Fig. 11.