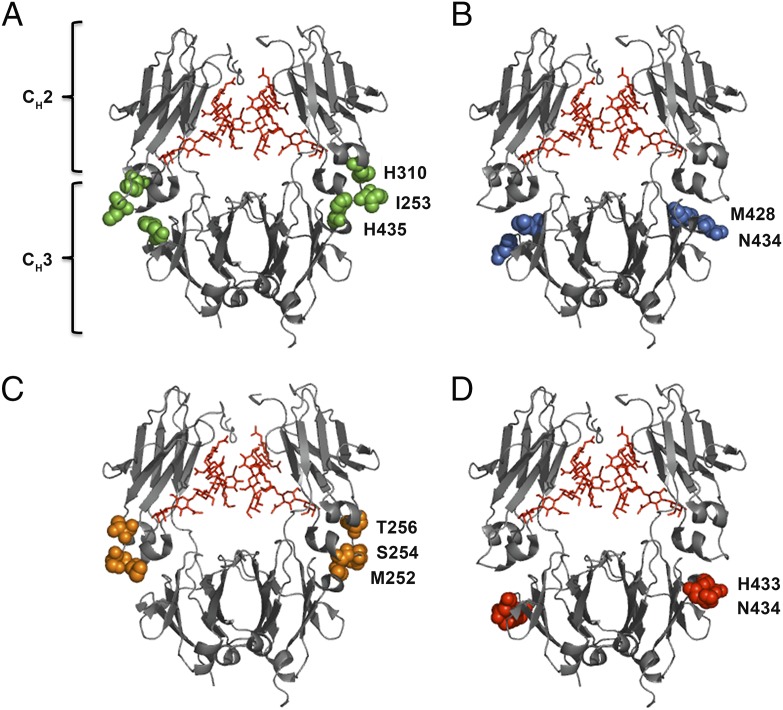
FIGURE 1.
Crystal structure illustrations of hIgG1 Fc variants. Amino acid residues targeted to modulate binding to hFcRn are highlighted. (A) The key Fc residues involved in binding to FcRn (I253, H310, and H435) at the CH2–CH3 interface are highlighted in green spheres. (B) M428 and N434 of the CH3 domains are highlighted in blue spheres. (C) The Fc residues M252, S254, and T256 of the CH2 domains are highlighted in orange spheres. (D) H433 and N434 of the CH3 domains are highlighted in red spheres. In all crystal structure illustrations the biantennary glycans attached to the N297 residues of the CH2 domains are shown in red. The figures were designed using PyMOL (http://www.pymol.org) with the crystallographic data of hIgG1 (Protein Data Bank accession code 1HZH) (70).