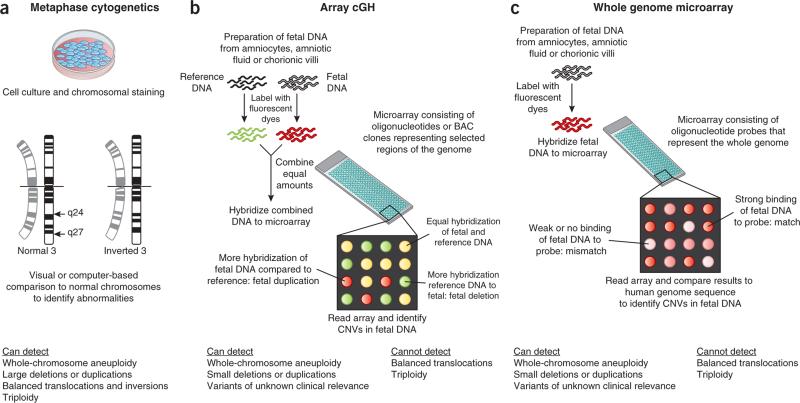
Figure 1.
Outline of the three major current techniques for analyzing fetal chromosomes. (a) Fetal cells are cultured and analyzed during cell division in metaphase. Chromosomes are analyzed under the microscope for the presence of dark and light staining bands. The staining patterns are compared with normal reference standards. Only relatively large deviations from normal (~5–10 Mb) can be detected. (b,c) The DNA within the fetal chromosomes, rather than the fetal chromosome itself, is compared to reference genomes. The DNA can be isolated from fetal cells or cell-free amniotic fluid with or without prior cell culture. In array comparative genomic hybridization (cGH) (b), patient and reference DNA samples are labeled with competing fluorescent dyes and hybridized to an array that contains DNA probes. Each probe is known to map to a specific region of the human genome. When the array is read, areas of mismatch appear as red or green. Special software converts the signal to indicate the affected area of the genome. In the method shown in c, only the patient's DNA is hybridized to an array that contains oligonucleotides (~60 bp) with coverage across the human genome. Areas of mismatch between the patient's DNA and the reference sequence are identified as CNVs. BAC, bacterial artificial chromosome.