Figure 4.
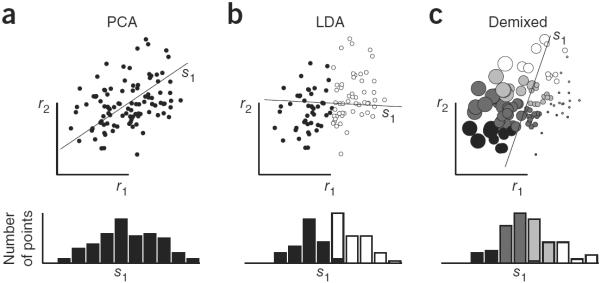
Conceptual illustration of PCA, LDA and demixed dimensionality reduction for two neurons (D = 2). (a) PCA finds the direction (s1 axis) that captures the greatest variance in the data (black dots, top), shown by the projection onto the s1 axis (bottom). (b) LDA finds the direction (s1 axis) that best separates the two groups of points (black and while dots, top). The separation can be seen in the projection onto the s1 axis (bottom). (c) Demixed dimensionality reduction (using the method described in ref. 16 finds the direction that explains the variance in dot color (s1 axis, top) and an orthogonal direction (s2 axis, not shown) that explains the variance in dot size. The organization in dot color can be seen in the projection onto the s1 axis (bottom). Note that these illustrations were created using the same data points (dots), and it is the use of different methods (which exploit different data features, such as group membership in (b) or color and size in (c)) that produce different directions s1 across the top panels and different projections across the bottom panels.
