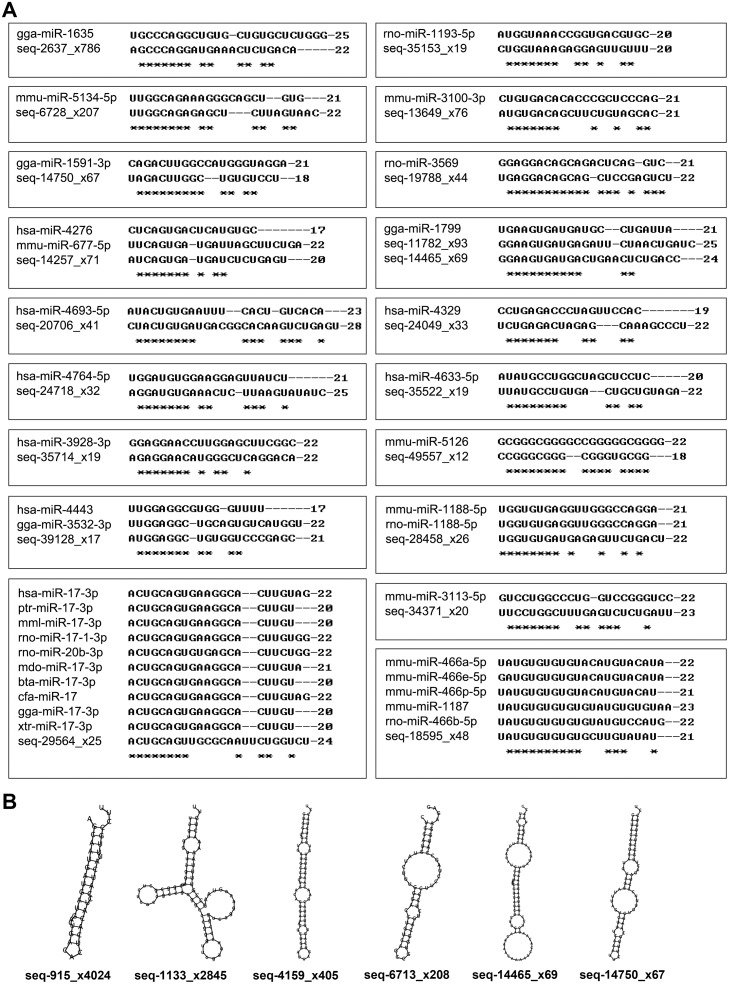
Fig 1. Novel miRNA candidates.
(a) Sequence alignment of novel miRNA candidates with known miRNAs of other species. *: conserved nucleotide. bta: Bos taurus. cfa: Canis familiaris. gga: Gallus gallus. hsa: Homo sapiens. mdo: Monodelphis domestica. mml: Macaca mulatta. mmu: Mus musculus. ptr: Pan troglodytes. rno: Rattus norvegicus. xtr: Xenopus tropicalis. (b) Secondary structures of putative precursor hairpins corresponding to six novel miRNA candidates identified in this study. One (seq-6713_x208) of these novel miRNAs were found to be up-regulated in late-gestational fetal KCs, whereas the other five miRNAs were down-regulated (see also S7 Table).