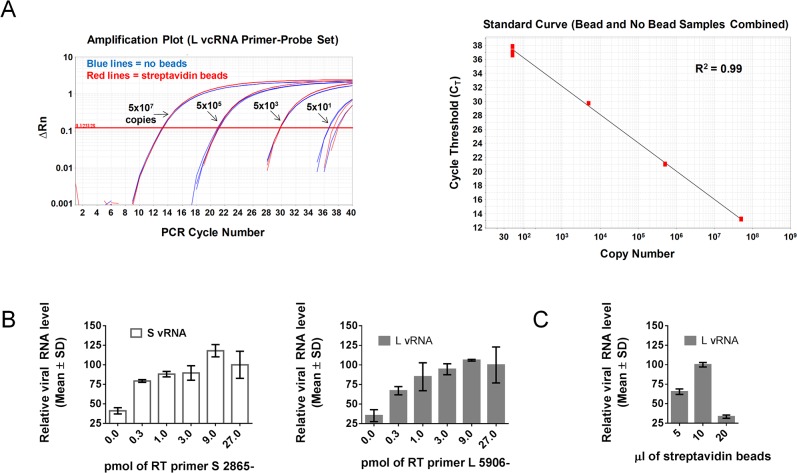
Fig 6. Optimization of assay conditions.
(A) Streptavidin beads do not impact QPCR efficiency. QPCR featuring the primer-probe set specific for LCMV L segment vcRNA listed in Table 3 was conducted using the pT7 plasmid encoding the LCMV L segment. The primer-probe set was tested on a range of plasmid template quantities (50, 5x103, 5x105, or 5x107 copies) with either 25 μl of streptavidin beads (red lines in the amplification plot) or no beads (blue lines in the amplification plot) included in the QPCR reaction mixture. The standard curve fit line was generated by pooling all of the values from samples containing beads or not. Although not shown, similar results were obtained for the remaining primer-probe sets specific for S vRNA and vcRNA or L vRNA. (B) 9 pmol of primer is sufficient to prime total viral RNA template for cDNA synthesis during RT. RNA extracted from sucrose-banded LCMV virions collected from Vero E6 cells at 48 hr pi was subjected to RT using 27, 9, 3, 0.3, or 0 pmol of biotinylated primer S 2865- (to target S vRNA) or L 5906- (to target L vRNA), followed by QPCR using the primer-probe sets for S vRNA or L vRNA, respectively, that are listed in Table 3. Data are presented as mean ± SD relative to the 9 pmol samples. (C) 10 μl of streptavidin beads is sufficient to capture 9 pmol of biotinylated RT primer. RNA extracted from sucrose-banded LCMV particles collected from Vero E6 cells at 48 hr pi was subjected to RT using 9 pmol of biotinylated RT primer S 5906- (specific for L vRNA). Following RT, biotinylated cDNAs were affinity purified using 20, 10, or 5 μl of magnetic streptavidin beads and subjected to QPCR using the primer-probe set for L vRNA listed in Table 3. Data are presented as mean ± SD relative to the 10 μl bead samples.