Fig 7. Use of optimized QRT-PCR assay to measure the RNA content in LCMV virions and the dynamics of genome replication during acute and persistent LCMV infection.
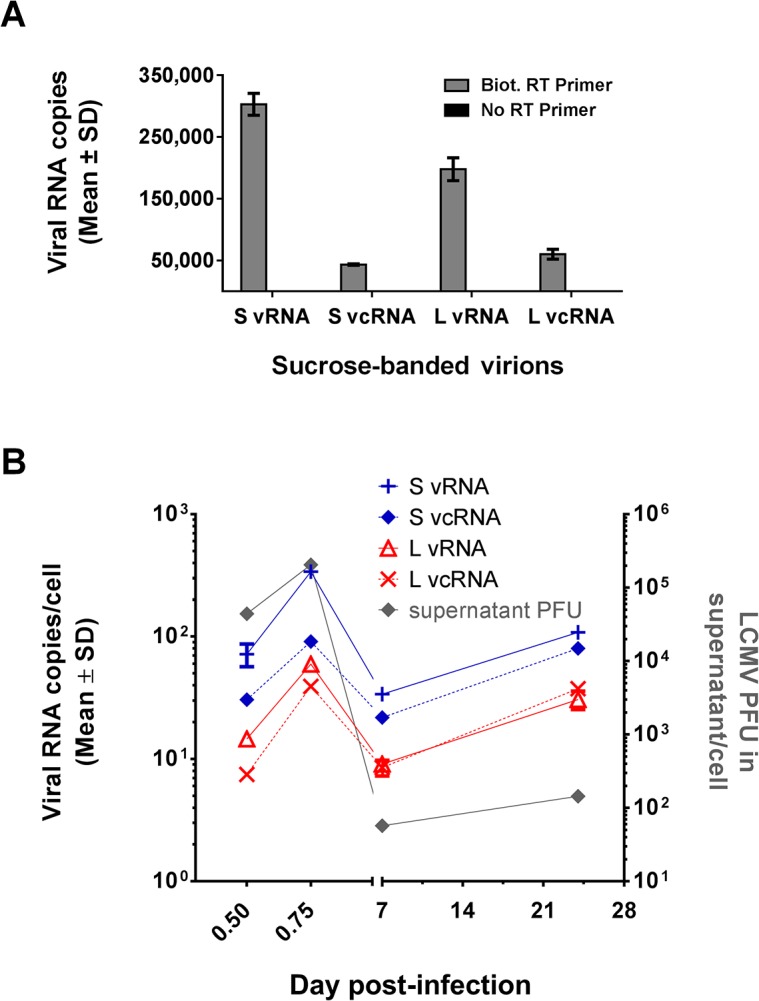
(A) Viral RNA content of LCMV virions. RNA extracted from sucrose-banded LCMV particles collected from Vero E6 cells at 48 hr pi was subjected to RT using 9 pmol of the biotinylated RT primers listed in Table 1 (gray bars). As a control, RT was also carried out in the absence of an RT primer (black bars). Biotinylated cDNAs were affinity purified using 10 μl of magnetic streptavidin beads and subjected to QPCR using the primer-probe sets for the vRNAs or vcRNAs of the L or S segment that are listed in Table 3. Data are presented as mean ± SD. (B) Dynamics of LCMV replication during acute and persistent LCMV infection. Viral RNA extracted from MC57 cells at 12 h, 18 hr, 7 d, or 24 d pi with LCMV was subjected to RT using 9 pmol of the biotinylated RT primers listed in Table 1. Biotinylated cDNAs were affinity purified using 10 μl of magnetic streptavidin beads and subjected to QPCR using the primer-probe sets for S vRNA or L vRNA, respectively, that are listed in Table 3. Values are listed as mean copies per cell ± SD and are plotted on the left Y axis. For each sample, PFUs in the supernatants were determined by plaque assay and are reported as PFU/cell on the right Y axis.
