Abstract
Transition of an evolving population to a new adaptive optimum is predicted to leave a signature in the distribution of effect sizes of fixed mutations. If they affect many traits (are pleiotropic), large effect mutations should contribute more when a population evolves to a farther adaptive peak than to a nearer peak. We tested this prediction in wild threespine stickleback fish (Gasterosteus aculeatus) by comparing the estimated frequency of large effect genetic changes underlying evolution as the same ancestor adapted to two lake types since the end of the ice age. A higher frequency of large effect genetic changes (quantitative trait loci) contributed to adaptive evolution in populations that adapted to lakes representing a more distant optimum than to lakes in which the optimum phenotype was nearer to the ancestral state. Our results also indicate that pleiotropy, not just optimum overshoot, contributes to this difference. These results suggest that a series of adaptive improvements to a new environment leaves a detectable mark in the genome of wild populations. Although not all assumptions of the theory are likely met in natural systems, the prediction may be robust enough to the complexities of natural environments to be useful when forecasting adaptive responses to large environmental changes.
Keywords: Adaptation, adaptive peak shift, natural selection, pleiotropy, QTL
Understanding how natural selection has produced the remarkable fits we observe between organisms and their environment is an important task for evolutionary biologists. Although natural selection acts on phenotypic traits, evolutionary response is determined by the genes that control these traits. Yet, we lack answers to some of the most basic questions about the genetics of adaptation. For example, the number of genes involved in adaptation to a new environment and their magnitude of effects remain largely unknown (Orr 1998; Phillips 2005, Barrett et al. 2006; Hoekstra and Coyne 2007; Stinchcombe and Hoekstra 2008; Linnen et al. 2009, Linnen and Hoekstra 2010).
According to Fisher’s geometric model of adaptation, genetic changes of large effect are expected to contribute infrequently to adaptation in new or changing environments (Fisher 1930, reviewed in Orr 2005). This follows from Fisher’s assumption that mutations likely affect many traits (i.e., are pleiotropic), with the result that large effect changes are more likely than small-effect changes to direct the population away from, rather than toward, a local adaptive peak (Fisher 1930). However, theory and laboratory experiments show that this disadvantage of large effect mutations is lessened when a population is far from the optimal phenotype (Orr 1998; Burch and Chao 1999; Orr 1999; Wahl and Krakauer 2000; Griswold and Whitlock 2003; Barrett et al. 2006; Kassen 2009; Gifford et al 2011) (Fig. 1). Whether this prediction is met in natural populations following an adaptive peak shift remains untested.
Figure 1.
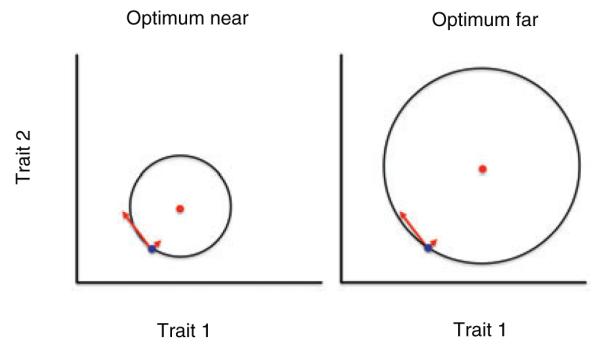
Illustration of Fisher’s geometric model of adaptation, in which the trait means of the same ancestral population are indicated by a blue dot in both panels. Arrows indicate two mutations, one of large effect (long arrow) and the other of small effect (short arrow). In the right panel the ancestral population begins the process of adaptation to a distant optimum (red dot); in the left panel the optimum (red dot) is nearer to the same ancestral population. Black circles are contours of equal fitness, with fitness higher inside the circle than outside. A small effect mutation is just as likely to increase fitness in both cases, but a large effect mutation is more likely to increase fitness when the optimum is far (right panel) than when it is near (left panel); this difference is enhanced with greater numbers of traits.
Figure 1. Illustration of Fisher’s geometric model of adaptation, in which the trait means of the same ancestral population are indicated by a blue dot in both panels. Arrows indicate two mutations, one of large effect (long arrow) and the other of small effect (short arrow). In the right panel the ancestral population begins the process of adaptation to a distant optimum (red dot); in the left panel the optimum (red dot) is nearer to the same ancestral population. Black circles are contours of equal fitness, with fitness higher inside the circle than outside. A small effect mutation is just as likely to increase fitness in both cases, but a large effect mutation is more likely to increase fitness when the optimum is far (right panel) than when it is near (left panel); this difference is enhanced with greater numbers of traits.
A difficulty when testing the prediction in nature is the inability to ensure that all assumptions of the geometric model of adaptation are strictly met. For example, the theory assumes that all adaptation occurs from new mutations (Fisher 1930; Orr 1998), whereas adaptation from standing genetic variation is common (Barrett and Schluter 2008; Renault et al. 2011). A test also requires appropriate controls and replication, which are difficult to verify in wild populations. Nevertheless, predictions concerning the frequency of large effect mutations may be relatively robust because unlike small effect mutations the probabilities of fixation of large effect mutations from standing variation and new mutation are similar (Hermisson and Pennings 2005). Despite potential limitations, tests in wild populations are vital to determining whether the theory is of any use in explaining patterns of phenotypic evolution.
We used threespine stickleback (Gasterosteus aculeatus) to test the prediction that adaptation to a phenotypic optimum farther from the ancestral state, compared with an optimum nearer to the ancestral state, will involve the fixation of a higher frequency of relatively large effect genetic changes (Orr 1998). The threespine stickleback is an ideal system because the same ancestral marine species has colonized and adapted independently to many new freshwater environments across the northern hemisphere since the last ice age, within the last 20,000 years. In freshwater, stickleback populations have repeatedly evolved reduced bony defensive armor and become bulkier and less streamlined in body shape compared to the marine ancestor (Taylor and McPhail 1986; Bell et al. 1993; Walker 1997). Other phenotypic changes have also evolved in parallel, such as spine position, pelvic girdle reduction, bone shapes, and rotation of jaw elements (Bell et al. 1993; Schluter and McPhail 1993; Walker and Bell 2000; Albert et al. 2008). Marine and freshwater populations can be crossed to produce fertile hybrids, permitting the genetic study of species differences (Peichel et al. 2001). Three genes of large effect that underlie adaptive changes in specific traits in freshwater have already been identified: Ectodysplasin, controlling bony lateral plate armor (Colosimo et al. 2005), Pitx1, controlling presence of the pelvic girdle (Shapiro et al. 2004; Chan et al. 2010), and Kitlg, controlling pigmentation (Miller et al. 2007). However, the general circumstances under which large effect genetic changes contribute to adaptation are unknown.
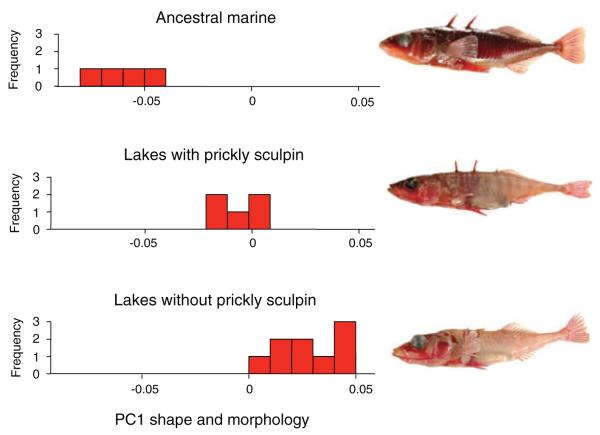
We compared the magnitude of quantitative trait loci (QTL) effect sizes underlying shape and armor adaptation in four freshwater populations residing in two contrasting lake types from coastal British Columbia, Canada, all of which formed approximately simultaneously about 12,000 years ago (Hutchinson et al. 2004). In two of these lakes, the prickly sculpin (Cottus asper) is present, an intraguild predatory fish that preys upon stickleback and consumes benthic invertebrate resources (Vamosi 2003; Ingrim et al. 2012). The remaining two lakes are physically similar but prickly sculpin are absent. On average, stickleback inhabiting lakes with prickly sculpin have intermediate armor and body shape compared to the marine ancestral population (which is more heavily armored and streamlined) and to stickleback adapted to non-sculpin lakes (which are bulkier and have less armor) (Fig. 2; Ingram et al., 2012). The consistency of these phenotypic differences between stickleback evolving in lakes with and without sculpin suggest that the two lake types represent distinct phenotypic optima for stickleback in freshwater, with lakes inhabited by sculpin having a phenotypic optimum for stickleback closer to the ancestral state than the optimum in non-sculpin lakes.
Figure 2.
Fish found in the ocean (top panel) and two contrasting freshwater lake types having distinct phenotypic optima for stickleback: lakes with the predatory and competitive sculpin, such as Paq and Graham Lakes (middle); and lakes that lack sculpin, such as Cranby and Hoggan Lakes (bottom). Data from Ingram et al. (2012) were used to measure the 54 landmark traits and one metric trait, pelvic spine length, on 25 specimens from freshwater stickleback populations in lakes with (n= 5 lakes) and without (n= 9) prickly sculpin. Three marine populations were also sampled, one of which was represented by two separate samples, one from the wild and the other from a population reared in freshwater ponds on the campus of the University of British Columbia (Pritchard and Schluter 2001). The first principal component accounted for over 75% of the variation among all population means. Analysis of traits was carried out in R 2.6.0.
Figure 2. Fish found in the ocean (top panel) and two contrasting freshwater lake types having distinct phenotypic optima for stickleback: lakes with the predatory and competitive sculpin, such as Paq and Graham Lakes (middle); and lakes that lack sculpin, such as Cranby and Hoggan Lakes (bottom). Data from Ingram et al. (2012) were used to measure the 54 landmark traits and one metric trait, pelvic spine length, on 25 specimens from freshwater stickleback populations in lakes with (n= 5 lakes) and without (n= 9) prickly sculpin. Three marine populations were also sampled, one of which was represented by two separate samples, one from the wild and the other from a population reared in freshwater ponds on the campus of the University of British Columbia (Pritchard and Schluter 2001). The first principal component accounted for over 75% of the variation among all population means. Analysis of traits was carried out in R 2.6.0.
We crossed ancestral marine stickleback with fish from sculpin-present and sculpin-absent lakes to compare the frequency of large effect QTL underlying shape and armor adaptation in populations residing in lakes of the two types. The advantage of a QTL approach is that it allows estimation of effect sizes of anonymous genes across the genome for wild populations. Although QTL are not necessarily single genes or mutations, they are a reasonable substitute in stickleback according to previous mapping, sequencing, and transgenic experiments on the genetic factors controlling stickleback traits in crosses of similar size, which have implicated one or a small number of genes within mapped QTL regions (Shapiro et al. 2004; Colosimo et al. 2005; Miller et al. 2007; Chan et al. 2010). The number of unique mutations and fixation events producing the QTL effects detected here cannot yet be determined for these populations and remains a problem for further study. Nevertheless, the absence of a difference in the frequency of large effect QTL between populations adapting to the two lake types would be sufficient grounds to conclude that the theoretical prediction is not met. We focused on shape and armor because they represent a suite of adaptive traits evolving together in a similar direction from the ancestral marine state, and because they are measured in the same units, permitting comparison of genetic effect sizes.
Methods
SAMPLING AND EXPERIMENTAL CROSSES
All study populations were from southwestern British Columbia, Canada. We used the anadromous marine population from the mouth of the Little Campbell River (49°1′4”, 122°45′52”). The two populations occurring with prickly sculpin were from Paq Lake (49°36′45”, 124°01′20”) and Graham Lake (49°31′00”, 124°45′00”). The two populations occurring without prickly sculpin were Cranby Lake (49°42′00”, 124°30′00”) and Hoggan Lake (49°09′00”, 123°50′00”). Adult sticklebacks were collected from each population using minnow traps in 2001 and brought to the laboratory.
Our breeding design involved artificially crossing a grand male from each freshwater lake with a grand female from the marine population to generate F1 marine-by-freshwater crosses for each lake. This cross design maximizes the number of offspring (due to the higher average fecundity of marine females), and generates F1 progeny carrying the marine mitochondrial genome, the freshwater X or Y-chromosome, and both marine and freshwater variants at all other loci. Use of a single individual from each population is sufficient to include all fixed autosomal differences between marine and freshwater fish, with nonfixed differences represented with probability proportional to frequency. In 2002, we artificially crossed males and females from the same F1 families to generate four F2 crosses for each lake (Cranby Lake, total N= 374; Paq Lake N= 374; Hoggan Lake N= 290; and Graham Lake N= 361). All fish were raised in 27-gallon aquaria in a single room held at 18°C with 16 h of light per day and fed brine shrimp and frozen blood worms to satiation daily. F2 progeny were raised to maturity with a minimum standard length of 40 mm before being euthanized with an overdose of MS-222 and preserved in 95% EtOH. The caudal fin was removed for DNA extraction.
MORPHOLOGICAL ANALYSIS
Bony armor of preserved stickleback was stained by soaking all specimens for 24–48 h in a 1% KOH solution containing alizarin red followed by preservation in a 40% isopropyl alcohol solution. Digital photographs of the right side of each fish were taken with a Nikon D1H camera. To quantify shape differences between populations, we placed 27 landmarks on each photo using tpsDig (Rohlf 2005), as described (Albert et al. 2008) (Fig. S1). To ensure consistency of shape coordinates among populations, landmark data were aligned and corrected for geometric size using the mean Cranby orientation as our reference and rotating the landmark configurations from other populations to correspond to this reference with the SHAPES library in R 2.6.0 (R Development Team 2005). This resulted in 27 x and 27 y coordinates defining the 54 landmark traits (Table S1).
Ethanol preservation causes bending of specimens in the vertical direction, which contributes to significant shape variation. We removed this bending effect from the 54 aligned x and y coordinates of the F2 progeny with principal components analysis, as described (Albert et al. 2008). The first eigenvector (principal component, PC1) explained 37% of the variation and described a U-shaped displacement upward of the most anterior and posterior landmarks and a simultaneous downward displacement of landmarks near the center of the body, consistent with the observed bending of preserved specimens. PC1 failed to map to any QTL location, consistent with previous findings and suggesting the bending was associated with measurement error (Albert et al. 2008). We therefore deleted PC1 and the first eigenvector from loadings to remove this source of landmark variation. The adjusted shape coordinates were obtained by back-transforming the PC scores to the original 54 x and y coordinates. These error-corrected x and y coordinates were used as the phenotypes for all subsequent QTL analyses. We measured an additional 10 metric traits known to differentiate marine and freshwater populations, taking the residual values from a regression line having common slope to adjust for differences in standard body length. We did not include number, length, or width of lateral plates (Colosimo et al. 2005), because these armor traits consistently have a similar mean in lakes with and without sculpin and show no evidence of an intermediate optimum in sculpin-present lakes. Altogether, 64 morphological traits commonly associated with adaptation to freshwater were measured for QTL analyses (Table S1, Fig. S1).
GENETIC ANALYSIS
We isolated total genomic DNA from caudal fin clips using a standard proteinase K phenol chloroform protocol (Sambrook et al. 1989). We quantified DNA yield using spectrophotometry, standardizing all samples to 5ng/μl, and then preserving these samples at −20°C. Microsatellites were amplified by polymerase chain reaction (PCR) with a 384-well DNA Engine® Peltier Thermal Cycler (Bio-Rad, Inc., Hercules, CA) in 5 μl reactions containing 5 ng of genomic DNA, 0.5 μM of each forward and reverse primer (Table S2), 1 × PCR buffer, 0.125 mM of each dNTP, 1.5 mM MgCl2, and 0.125U of AmpliTaq Gold polymerase (Applied Biosystems, Foster City, CA). Cycling conditions were standardized over all loci as follows: 93°C for 3 min, 95°C 30 s, 59°C 30 s, 72°C 30 s, 5 cycles of 94°C 30 s, 59°C 30 s, 72°C 30 s, 35 cycles of 90°C 30 s, 60°C 30 s, 72°C 30 s, followed by 72°C for 10 min and then cooled to 4°C. Electrophoresis consisted of pooling PCR products with an internal size standard (LIZ 500 bp, Applied Biosystems) and loading onto the Applied Biosystems 3730S Automated Sequencer. Allelic sizes (in base pairs) were determined by reference to the internal sizing standard in the software GENEMAPPER version 3.7 (Applied Biosystems).
We screened over 250 microsatellite loci in wild fish, grandparents, and F1 of each population, selecting the most informative microsatellites for phylogenetic analysis and genetic mapping. A total of 104 microsatellites were genotyped in wild marine and freshwater populations to quantify the genetic distance between marine and freshwater populations. We used the chord genetic distance (Cavalli-Sforza and Edwards 1967), as postglacial mutation has been shown to be an insignificant factor in differences in allelic frequency variation between marine and freshwater stickleback populations (Taylor and McPhail 2000), and assessed the relative demographic history of the populations by comparing δμ2 estimates following Goldstein et al. (1999). These data also provided an indication whether the rate of fixed differences or expected heterozygosity estimates differed at these markers among freshwater environments. Genetic diversity estimates were calculated with ARLEQUIN version 3.0 (Excoffier et al. 2005), while phylogenetic relationships among populations were estimated by bootstrapping over loci in POPULATIONS version 1.2.28 (Langella 2002).
LINKAGE MAPPING AND QTL ANALYSES
A total of 96 microsatellites were genotyped in the Cranby F2, 94 in the Paq F2, 75 in the Hoggan F2, and 85 in the Graham F2 to assemble linkage maps for each population, including a diagnostic marker for sex (isocitrate dehydrogenase, Peichel et al. 2004) (Table S2). The number and selection of markers in each population were designed to minimize redundancy and maximize genome coverage. We chose markers at intervals of approximately 15 centiMorgans (cM), sufficient for the size of genomic regions expected to segregate in an F2 cross of the size used here (Broman 2001). Marker sets were not identical between populations, but their coverage of regions was comparable. Linkage maps for each cross were generated with JOINMAP 3.0 (Van Ooijen and Voorrips 2001). We created a locus data file for each of the four F2 families generated for each lake. We set the segregation type to cross-pollinator (CP), which allows for up to four alleles per locus, appropriate for outbred crosses from natural populations. We grouped microsatellite loci into linkage groups with a log of odds ratio (LOD) score threshold of 4.0 and created a map using the Kosambi mapping function with a LOD threshold of 3 (P = 0.001), a recombination threshold of 0.499, a jump threshold of 5.0, and a triplet value of 5.0. In the rare event that orders of markers along a linkage group differed between families, we compared the data to previously established meiotic linkage groups. In these cases, the most commonly observed order was deemed the most likely and this fixed order was applied subsequently. For each population, we combined the data from the separate families to calculate an integrated map under the JOIN menu, which uses mean recombination fractions and combined LOD scores. Chance recombination events and biological variation can nevertheless generate differences between populations in estimates of map distances. However, because we combined information from different families within the same population, our maps are robust for directly comparing QTL at similar locations between families.
We scanned both maps for QTL using the Haley–Knott (HK) regression method (Haley and Knott 1992) implemented in R/qtl 2 with the scanqtl function (Broman et al. 2003). Genotype and phenotype data files were created with the four-way cross format (allowing for four alleles segregating in the F2) and imported with the read.cross function. Genotype probabilities, needed for interval mapping, were estimated with the calc.geneprob function with a step size of 2 cM. We used sex as a covariate in the HK analysis to control for sexual dimorphism in traits (Albert et al. 2008). A population QTL map was obtained by summing the LOD scores of separate QTL maps of its four F2 families. We used a LOD threshold of 7.1 for QTL detection, since our aim was to compare effect size distributions rather than to test individual QTL. This threshold corresponds to a tail probability of 0.001 under a chi-square distribution having 12 degrees of freedom. This is the same tail probability as that corresponding to a LOD threshold of 3.5 in a single cross of equivalent size (3 df), and corresponded to a tail probability of approximately 0.10 in genome-wide permutation tests.
Effect sizes and percent variance explained (PVE) for significant population QTL were estimated by fitting a linear model including all significant QTL affecting a given trait with the fitqtl function in R/qtl using 128 imputations and averaged over the four F2 families. Under this model, the PVE (as well as the F-values and P-values) are approximations from the LOD score. Effect size of a QTL for a given trait was estimated as the difference between the mean of F2 genotypes homozygous for the marine alleles and that homozygous for the freshwater alleles at the QTL. This difference was scored as a positive effect if the mean of the freshwater F2 genotype at the QTL was closer to the mean of the freshwater population than was the mean of the marine genotype (Table S1). Otherwise, the direction of QTL effect was scored as negative. We converted all QTL effect sizes to millimeters by multiplying all of them by a constant calculated as the mean standard length of all the fish divided by the mean distance between the most anterior and posterior landmark coordinates (x1, y1 and x17, y17). This conversion does not alter relative sizes of landmark configurations. We divided the effect size of metric QTL by the square root of two to account for the greater expected average distance between a pair of landmarks in two-dimensional metric traits (x and y) compared with a position change in a one-dimensional shape trait (x or y). The sign of PVE was also fixed to match the direction of the corresponding QTL effect size.
Results
Phylogenetic and population genetic analyses of the marine and freshwater stickleback supported geological evidence that freshwater populations arose independently and approximately simultaneously from the marine population (chord distance: Marine-Paq = 0.61, Marine-Cranby = 0.61, Marine-Graham = 0.61, Marine-Hoggan = 0.62, Fst: Marine-Paq = 0.12, Marine-Cranby = 0.11, Marine-Graham = 0.12, Marine-Hoggan = 0.14) (Fig. S2). Stepwise mutation models were also consistent with the observation that freshwater populations were similar in relative phylogenetic age from the marine ancestor (δμ2: Marine-Paq = 0.29, Marine-Cranby = 0.31, Marine-Graham = 0.20, Marine-Hoggan = 0.15).
Four genetic linkage maps, two from each lake type, were assembled from the microsatellite data (Fig. S3). We achieved complete genome coverage over the 21 linkage groups known for threespine stickleback with an average total map length of 993 cM and a marker density of one microsatellite every 15 cM (average interval size in cM between loci; Cranby = 14.9, Hoggan = 14.7 cM, Paq = 16.0, Graham = 14.0, Fig. S3, Table S2). Although chance recombination events or biological variation generated minor differences among maps, total map lengths were highly similar to previously generated marine x freshwater stickleback maps (Peichel et al. 2001; Albert et al. 2008). We detected multiple QTL for landmark and metric traits in all F2 crosses. The x and y coordinates of individual landmarks always mapped to different QTL, justifying treating them as separate traits (Albert et al. 2008). We found 36 QTL mapping to 12 linkage groups in the crosses with fish from sculpin-present lakes. We detected 41 QTL mapping to 13 linkage groups in the crosses with fish from sculpin-absent lakes (Table 1).
Table 1.
Location and effect size of significant QTL for landmark (x, y), and size-adjusted metric traits (s28–s37). See Figure S1 and Table S1 for description of traits. Lg refers to linkage group, position refers to the most likely QTL location in centiMorgans, LOD is the log of odds ratio, and PVE is the percentage of phenotypic variance explained. Effect size and direction (positive and negative) of QTL refers to the magnitude of change (in millimeters) when replacing two marine alleles with freshwater alleles at the QTL.
| Population | Trait | Lg | Position | LOD | PVE | Effect size and direction |
|---|---|---|---|---|---|---|
| Cranby | s37 | 1 | 50 | 9.28 | 7.21 | 0.0035 |
| Cranby | x4 | 1 | 50 | 10.63 | 12.31 | 0.3357 |
| Cranby | x23 | 2 | 6 | 8.15 | 9.48 | −0.0820 |
| Cranby | x26 | 4 | 44 | 8.95 | 9.06 | 0.1611 |
| Cranby | y4 | 4 | 44 | 8.26 | 8.52 | 0.0939 |
| Cranby | s32 | 4 | 46 | 13.99 | 13.98 | 0.3678 |
| Cranby | x3 | 4 | 46 | 9.21 | 7.15 | −0.2971 |
| Cranby | s33 | 4 | 48 | 35.38 | 28.87 | 1.0321 |
| Cranby | s35 | 4 | 50 | 14.50 | 15.41 | −0.2750 |
| Cranby | x8 | 4 | 50 | 10.98 | 11.16 | 0.3682 |
| Cranby | y5 | 4 | 50 | 17.62 | 18.35 | 0.5192 |
| Cranby | s34 | 4 | 50 | 46.90 | 40.56 | 1.0315 |
| Cranby | s28 | 4 | 52 | 12.76 | 13.57 | 0.7285 |
| Cranby | x24 | 4 | 54 | 13.98 | 14.68 | 0.3185 |
| Cranby | y2 | 7 | 30 | 7.78 | 8.90 | −0.1388 |
| Cranby | s31 | 7 | 34 | 9.64 | 6.87 | 0.0105 |
| Cranby | y8 | 7 | 45 | 8.58 | 9.40 | 0.1117 |
| Cranby | s36 | 8 | 22 | 7.77 | 8.85 | −0.0806 |
| Cranby | y6 | 9 | 16 | 7.83 | 8.62 | 0.0786 |
| Cranby | y12 | 12 | 30 | 8.12 | 8.95 | 0.0159 |
| Cranby | x12 | 12 | 36 | 9.74 | 9.60 | 0.1123 |
| Cranby | x11 | 12 | 40 | 7.84 | 7.57 | 0.0903 |
| Cranby | x17 | 16 | 27 | 7.75 | 6.34 | −0.0948 |
| Cranby | x1 | 18 | 0 | 9.60 | 10.45 | −0.2108 |
| Cranby | s37 | 18 | 4 | 12.88 | 10.61 | −0.1786 |
| Cranby | y25 | 21 | 0 | 10.31 | 10.61 | −0.0221 |
| Cranby | y26 | 21 | 0 | 10.03 | 9.97 | −0.0136 |
| Hoggan | x12 | 1 | 0 | 8.10 | 9.07 | −0.0951 |
| Hoggan | x17 | 1 | 0 | 8.63 | 6.44 | −0.0835 |
| Hoggan | s32 | 4 | 38 | 11.27 | 15.12 | 0.2720 |
| Hoggan | s34 | 4 | 42 | 16.51 | 22.75 | 0.6804 |
| Hoggan | s33 | 4 | 42 | 12.92 | 15.74 | 0.9389 |
| Hoggan | y15 | 4 | 44 | 8.39 | 13.06 | 0.1208 |
| Hoggan | x6 | 4 | 44 | 8.85 | 11.81 | 0.3191 |
| Hoggan | s28 | 4 | 44 | 10.34 | 13.85 | 0.7743 |
| Hoggan | x25 | 7 | 34 | 9.77 | 12.83 | 0.1989 |
| Hoggan | x24 | 7 | 34 | 8.60 | 13.32 | 0.2654 |
| Hoggan | s29 | 12 | 14 | 9.21 | 10.30 | −0.1171 |
| Hoggan | s28 | 13 | 16 | 7.65 | 10.50 | −0.5081 |
| Hoggan | y9 | 19 | 16 | 7.80 | 9.79 | 0.0288 |
| Hoggan | s33 | 20 | 8 | 9.71 | 11.72 | 0.6897 |
| Graham | y22 | 1 | 30 | 7.86 | 8.57 | 0.0513 |
| Graham | x23 | 1 | 68 | 7.71 | 9.11 | 0.1412 |
| Graham | y5 | 4 | 50 | 8.48 | 10.29 | 0.3040 |
| Graham | s32 | 4 | 56 | 9.16 | 10.40 | 0.3018 |
| Graham | y7 | 4 | 56 | 7.83 | 8.31 | 0.1931 |
| Graham | s34 | 4 | 60 | 10.98 | 11.91 | 0.5130 |
| Graham | y24 | 4 | 60 | 7.65 | 9.02 | 0.1955 |
| Graham | x24 | 4 | 2 | 9.51 | 10.63 | 0.2380 |
| Graham | s33 | 4 | 64 | 8.11 | 8.24 | 0.3758 |
| Graham | s34 | 9 | 2 | 10.33 | 12.08 | 0.2020 |
| Graham | s30 | 9 | 6 | 12.94 | 10.98 | −0.2427 |
| Graham | s37 | 12 | 24 | 8.14 | 9.04 | −0.1224 |
| Graham | y6 | 12 | 26 | 8.84 | 10.97 | −0.2330 |
| Graham | y2 | 12 | 30 | 7.59 | 10.05 | −0.1757 |
| Graham | x26 | 12 | 32 | 8.33 | 9.15 | 0.2506 |
| Graham | x5 | 12 | 40 | 8.22 | 9.66 | 0.1528 |
| Graham | y8 | 12 | 50 | 10.32 | 9.29 | 0.1474 |
| Graham | s30 | 12 | 55 | 9.13 | 7.66 | 0.0067 |
| Graham | y13 | 14 | 40 | 8.01 | 7.74 | −0.1440 |
| Graham | y18 | 14 | 40 | 7.86 | 7.99 | 0.1699 |
| Graham | x25 | 17 | 32 | 8.01 | 9.49 | 0.2376 |
| Graham | y27 | 20 | 44 | 8.43 | 9.28 | −0.1581 |
| Paq | x7 | 1 | 22 | 8.44 | 7.67 | −0.1548 |
| Paq | x27 | 1 | 34 | 7.73 | 8.92 | −0.2084 |
| Paq | x10 | 1 | 56 | 9.49 | 7.33 | 0.1963 |
| Paq | x9 | 1 | 58 | 8.25 | 8.59 | 0.1568 |
| Paq | x6 | 4 | 11 | 8.12 | 5.81 | 0.0689 |
| Paq | s29 | 10 | 40 | 9.16 | 8.17 | −0.0346 |
| Paq | x16 | 11 | 22 | 8.68 | 8.23 | −0.1602 |
| Paq | x25 | 12 | 38 | 10.03 | 11.07 | −0.2152 |
| Paq | x6 | 12 | 44 | 9.52 | 7.72 | 0.2201 |
| Paq | y27 | 13 | 16 | 8.85 | 8.80 | 0.1025 |
| Paq | y6 | 13 | 36 | 9.54 | 10.67 | 0.2036 |
| Paq | s37 | 16 | 14 | 8.06 | 9.59 | 0.1512 |
| Paq | y24 | 16 | 14 | 8.05 | 8.60 | 0.1098 |
| Paq | x20 | 18 | 0 | 7.62 | 8.98 | 0.0077 |
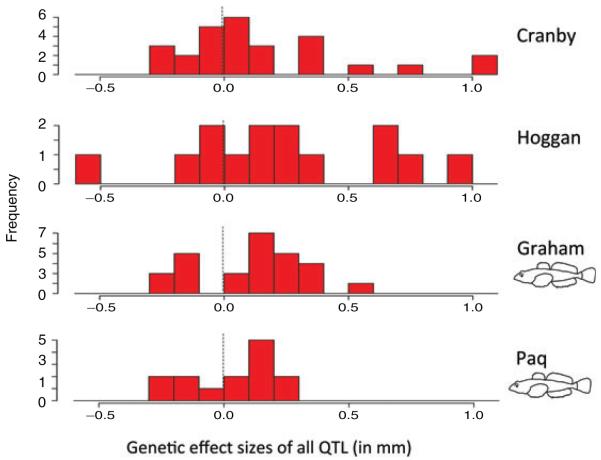
As expected, large effect genetic changes were relatively uncommon in both sculpin-present lakes (Paq and Graham), representing the near optimum type (Table 1), and sculpin-absent lakes (Cranby and Hoggan), representing the far optimum type (Table 1). Nevertheless, the frequency of large effect QTL was higher in stickleback adapting to the farther optimum, as judged by a higher standard deviation of both negative and positive genetic effect sizes when all traits are considered together (Fig. 3) (two sample t-test, one-sided t= 5.169, df = 2, P= 0.018). This difference was also significant when effect size was measured instead as percentage of variance explained (t= 3.25, df = 2, P= 0.042) (Fig. S4). The trend held for metric traits and landmark traits separately (Figs. S5, S6) but was statistically significant only for the metric traits (t= 4.406, df = 2, P= 0.024; landmark traits: t= 0.606, df = 2, P= 0.302). All of the largest effect genetic changes occurred in sculpin-absent lakes (e.g., the fraction of QTL with an effect size greater than the 95% quantile of all genetic effects: t= 7.667, df = 2, P= 0.008). Thus, the process of adaptation in stickleback took more large steps as they adapted to the more distant phenotypic optimum, beginning from the marine ancestral state (Fig. 3).
Figure 3.
The distribution and direction of QTL effect sizes for all traits measured in freshwater stickleback populations adapting to sculpin-absent lakes (Cranby and Hoggan) and sculpin-present lakes (Graham and Paq).
Figure 3. The distribution and direction of QTL effect sizes for all traits measured in freshwater stickleback populations adapting to sculpin-absent lakes (Cranby and Hoggan) and sculpin-present lakes (Graham and Paq).
Discussion
In this study we tested whether adaptation to a farther phenotypic optimum involved a higher frequency of large effect genetic changes than adaptation to a nearer optimum in wild populations of the threespine stickleback. Our test compared populations of approximately equal age that have adapted independently to two lake types, representing near and far morphological optima, beginning from the same ancestral state at the end of the last ice age. Previous studies of replicate populations showed that shape and armor of stickleback has evolved in a similar direction many times over in freshwater (e.g., Hagen and Gilberts 1973; Bell et al. 1993; Walker 1997; Walker and Bell 2000; Cresko et al. 2004; Schluter et al. 2004; Shapiro et al. 2004; Kimmel et al. 2005; Leinonen et al. 2006; Albert et al. 2008; Le Rouzic et al. 2011; Kaeuffer et al. 2012), but that stickleback in lakes with the intraguild predator, prickly sculpin, evolved to a mean phenotype closer to the ancestral state than stickleback in sculpin-absent lakes (Ingram et al. 2012). By crossing representative freshwater populations from the two lake types to the same marine population, we showed that populations in sculpin-absent lakes, representing the far-optimum environment, took more large steps than populations in sculpin-present lakes, representing the near optimum. As far as we know, this represents the first demonstration from wild populations of such a pattern, and is in agreement with theoretical expectations from the geometric model of adaptation. We recognize that this result is based on only four populations, two of each lake type, and a limited number of QTL overall, and that the generality of these findings therefore requires further investigation. Here we discuss the possible mechanisms underlying this pattern and some alternative hypotheses to explain it.
PLEIOTROPY VERSUS OPTIMUM OVERSHOOT
The results suggest that an important assumption of the theory is likely met in our study system. Under Fisher’s geometric model, deleterious side effects on other traits is the main reason that large effect mutations influencing a given trait are usually disadvantageous, an outcome that is predicted to become more severe the closer a population is to its adaptive optimum (Fisher 1930; Orr 1998). An alternative possibility is that large effect mutations are directly maladaptive in near-optimum populations because they cause single traits to overshoot the optimum and bring the population farther from it (Orr 1998). In our data, the largest average QTL effects we detected in far-optimum populations represented 0.55, 1.2, 1.6, and 2.8 times the total difference between the ancestral state and the mean of the near-optimum populations (numbers are for pelvic spine length, pelvic girdle length, ascending arch of the pelvic girdle [“y5”; Cranby Lake only], and ectocorocoid length; Tables 1, S1). This suggests that optimum overshoot may have contributed to the lower frequency of large effect QTL in near-optimum populations, but that it is not the only explanation. Except in the case of ectocorocoid length the large effect mutations from the far-optimum populations on their own would, in the near-optimum populations, have brought the ancestral populations closer to the near optimum.
While some large effect mutations may have low pleiotropy, likely increasing their chances of being advantageous (Fisher 1930), most genetic effects fixed by selection in these sticklebacks indeed appear to affect multiple traits. This is supported by the high frequency of negative effect QTLs, representing freshwater alleles whose effect on an individual trait is opposite to the direction of phenotypic evolution under selection (Fig. 3). It is predicted that when selection fixes pleiotropic mutations negative effects on individual traits occur but are compensated by positive effects on other traits, yielding a net positive effect on fitness. However, negative effects may also arise from stabilizing selection around the optimum if traits are polygenic (Griswold and Whitlock 2003). Several of the QTL we discovered demonstrate pleiotropic effects, including one on linkage group 4 responsible for the three largest QTL effects (top of ascending branch of the pelvic girdle, pelvic girdle length, and pelvic spine length) (Table 1). We do not yet know whether this represents a single pleiotropic mutation or multiple linked mutations. Finally, there are demonstrable pleiotropic effects of two previously identified large effect genes that underlie adaptive differences between marine and freshwater stickleback (Eda and Kitlg; Colosimo et al. 2005; Miller et al. 2007; Barrett et al. 2008; Harris et al. 2008). Altogether, these results suggest that pleiotropy is a common feature of adaptive evolution (Otto 2004) and may therefore have affected the distribution of effect sizes fixed during adaptation to freshwater environments in threespine stickleback. However, we remain cautious in this interpretation because we cannot rule out all other possibilities.
LIMITS TO TESTING THE THEORY IN NATURE
Our results support the prediction that selection should fix a greater proportion of large effect genetic changes when a population adapts to a more distant adaptive peak, but limitations and alternatives to the theory need to be considered in light of these findings. A simple possibility is that all stickleback populations are presently at a transient stage of a continuing process of adaptation to an unreached distant optimum. In this case those populations that happened to have experienced more large effect mutations will have attained a greater distance from the ancestral state by the present time. However, this possibility is unlikely because of the consistent association observed between mean phenotype and lake type (presence/absence of an intraguild predator) among independent populations that is the basis of our test.
Another alternative model is that all populations have adapted to an optimum phenotype that itself gradually moved further and further away from the ancestral state, and that different stopping points represent the different lake types occurring today. Perhaps sculpin were present in all the lakes at one time, for example, but then gradually disappeared from only some of them, shifting the adaptive peak further from the ancestor. Under this model, only small-effect mutations are advantageous, at least if the optima shift gradually. It also predicts that the distribution of genetic effect sizes should not differ between present-day environments (Kopp and Hermisson 2007), such as our different lake types. Our data do not agree with these expectations, but the model cannot be ruled out because we are as yet uncertain whether single QTL represent one fixation event or several. Perhaps, the effect sizes of the largest effect QTL are built up of multiple smaller-effect, linked mutations that fixed sequentially during adaptation (McGregor et al. 2007; Bickel et al. 2011). It may be possible to test this alternative by measuring effect sizes in populations of different ages at different stages in the process of adapting to a similar environment. We have not studied populations at intermediate stages of the adaptive process.
Existing theory for the geometric model assumes that adaptation occurs solely by the fixation of new mutations, whereas evolution from standing genetic variation is common in nature (Feder et al. 2003; Barrett and Schluter 2008; Renaut et al. 2011), including in stickleback (Colosimo et al. 2005; Miller et al 2007). The effect of adaptation from standing variation on the predicted distribution of effect sizes is not well studied and is beyond the scope of the current study. It is possible that the same qualitative predictions concerning the contribution of large effect mutations hold when adapting from standing genetic variation as when adaptation occurs from new mutations. This is because the fixation probability of large effect mutations is less affected by standing variation than is that of small effect mutations, which are likely to be lost when they first arise (Hermisson and Pennings 2005).
However, certain processes involving standing variation can increase the pool of large effect advantageous mutations. In the case of stickleback, we suspect (but do not yet have direct evidence) that standing variation for alleles advantageous in freshwater is maintained in the marine population by gene flow between marine and freshwater populations, which hybridize where they encounter one another in the breeding season (Colosimo et al 2005; McCairns and Bernatchez 2010). This beneficial variation is then available to marine colonists of new freshwater environments, speeding their rate of adaptation (Schluter and Conte 2009). The process would bias the standing pool of variation toward advantageous mutations, but the prediction that large effect alleles are more likely to be advantageous in far-optimum than in near-optimum freshwater environments should still hold. Nevertheless, depending on how long allele copies remain in the sea before being eliminated, closely linked freshwater alleles would remain linked for a time in the marine pool, increasing the chances that they would sweep to fixation together if present in freshwater colonists, behaving as a single large effect allele. Over time and across many populations, this process might allow the buildup and preservation of large effect alleles consisting of multiple closely linked advantageous mutations.
Conclusions
Fisher’s geometric model of adaptation reconciled Mendelian genetics with Darwinian gradualism, but conflict over the number and magnitude of the hypothesized genetic changes responsible for adaptation has persisted (Orr and Coyne 1992; Barton 1998; Orr 2005). Our finding of a higher frequency of large effect genetic changes in the population with a farther optimum may help to explain why genes of large effect are sometimes, but not always, found to contribute to adaptive differences between populations (Bradshaw et al. 1998; Burch and Chao 1999; Sawamura et al. 2000; Bernatchez et al. 2010). Adaptive peak shifts precipitated by rapid environmental change may be common in contemporary evolutionary scenarios. Despite potential limitations of current theory and the complexities of natural environments, these results demonstrate that it is essential to evaluate predictions of the geometric model in natural populations. To this end, our study demonstrates that theory can predict the circumstances under which genes of large effect might be advantageous in nature.
Supplementary Material
Acknowledgements
We thank R. Barrett, K. Marchinko, L. Harmon, P. Nosil, T. Vines, A. Albert, S. Vamosi, S. Vanderzwan, members of the Schluter Lab and Rogers Lab for comments and discussion, K. Faller, D. Yim, P. Louie, and J. Courchesne for assistance in the rearing and husbandry of stickleback. This research was supported by an Natural Sciences and Engineering Research Council of Canada (NSERC) Special Research Opportunity Grant to D. Schluter and grants from, NSERC Discovery, and the Alberta Innovates—Technology Futures New Faculty Award to SMR, and by an National Human Genome Research Institute Center of Excellence in Genomic Science Grant to DMK, who is an investigator of the Howard Hughes Medical Institute.
Literature Cited
- Albert AYK, Sawaya S, Vines TH, Knecht AK, Miller CT, Summers BR, Balabhadra S, Kingsley DM, Schluter D. The genetics of adaptive shape shift in stickleback: pleiotropy and effect size. Evolution. 2008;62(1):76–85. doi: 10.1111/j.1558-5646.2007.00259.x. [DOI] [PubMed] [Google Scholar]
- Barrett RDH, Schluter D. Adaptation from standing genetic variation. Trends Ecol. Evol. 2008;23(1):38–44. doi: 10.1016/j.tree.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Barrett RDH, Craig MacLean R, Bell G. Mutations of intermediate effect are responsible for adaptation in evolving Pseudomonas fluorescens populations. Biol. Lett. 2006;2(2):236–238. doi: 10.1098/rsbl.2006.0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett RDH, Rogers SM, Schluter D. Natural selection on a major armor gene in threespine stickleback. Science. 2008;322(5899):255–257. doi: 10.1126/science.1159978. [DOI] [PubMed] [Google Scholar]
- Barton NH. The geometry of adaptation. Nature. 1998;395:751–752. doi: 10.1038/27338. [DOI] [PubMed] [Google Scholar]
- Bell MA, Orti G, Walker JA, Koenings JP. Evolution of pelvic reduction in threespine stickleback fish—a test of competing hypotheses. Evolution. 1993;47(3):906–914. doi: 10.1111/j.1558-5646.1993.tb01243.x. [DOI] [PubMed] [Google Scholar]
- Bernatchez L, Renaut S. b., Whiteley AR, Derome N, Jeukens J, Landry L, Lu G, Nolte AW, òstbye K, Rogers SM, et al. On the origin of species: insights from the ecological genomics of lake whitefish. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2010;365:1783–1800. doi: 10.1098/rstb.2009.0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickel RD, Kopp A, Nuzhdin SV. Composite effects of polymorphisms near multiple regulatory elements create a major-effect QTL. PLoS Genet. 2011;7(1):e1001275. doi: 10.1371/journal.pgen.1001275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw H, Otto K, Frewen B, McKay J, Schemske D. Quantitative trait loci affecting differences in floral morphology between two species of monkeyflower (Mimulus) Genetics. 1998;149:367–382. doi: 10.1093/genetics/149.1.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K. Review of statistical methods for QTL mapping in experimental crosses. Lab Anim. 2001;30:44–52. [PubMed] [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- Burch CL, Chao L. Evolution by small steps and rugged landscapes in the RNA virus phi 6. Genetics. 1999;151:921–927. doi: 10.1093/genetics/151.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli LL, Edwards AWF. Phylogenetic analysis models and estimation procedures. Am. J. Hum. Genet. 1967;19:233. [PMC free article] [PubMed] [Google Scholar]
- Chan YF, Marks ME, Jones FC, Villarreal G, Jr., Shapiro MD, Brady SD, Southwick AM, Absher DM, Grimwood J, Schmutz J, et al. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a pitx1 enhancer. Science. 2010;327:302–305. doi: 10.1126/science.1182213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colosimo PF, Hosemann KE, Balabhadra S, Villareal G, Dickson M, Grimwood J, Schmutz J, Myers R, Schluter D, Kingsley DM. Widespread parallel evolution in sticklebacks by repeated fixation of ectodysplasin alleles. Science. 2005;307:1928–1933. doi: 10.1126/science.1107239. [DOI] [PubMed] [Google Scholar]
- Cresko WA, Amores A, Wilson C, Murphy J, Currey M, Phillips P, Bell MA, Kimmel CB, Postlethwait JH. Parallel genetic basis for repeated evolution of armor loss in Alaskan threespine stickleback populations. Proc. Natl. Acad. Sci. USA. 2004;101:6050–6055. doi: 10.1073/pnas.0308479101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol. Bioinform. Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Feder JL, Berlocher SH, Roethele JB, Dambroski H, Smith JJ, Perry WL, Gavrilovic V, Filchak KE, Rull J, Aluja M. Allopatric genetic origins for sympatric host-plant shifts and race formation in Rhagoletis. Proc. Natl. Acad. Sci. USA. 2003;100:10314–10319. doi: 10.1073/pnas.1730757100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. The genetical theory of natural selection. Oxford Univ. Press; Oxford , U.K.: 1930. [Google Scholar]
- Gifford D, Sijmen R, Schoustra E, Kassen R. The length of adaptive walks is insensitive to starting fitness in Aspergillus nidulans. Evolution. 2011;65(11):3070–3078. doi: 10.1111/j.1558-5646.2011.01380.x. [DOI] [PubMed] [Google Scholar]
- Goldstein DB, Roemer GW, Smith DA, Reich DE, Bergman A, Wayne RK. The use of microsatellite variation to infer population structure and demographic history in a natural model system. Genetics. 1999;151:797–801. doi: 10.1093/genetics/151.2.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold C, Whitlock M. The genetics of adaptation: the roles of pleiotropy, stabilizing selection and drift in shaping the distribution of bidirectional fixed mutational effects. Genetics. 2003;165:2181–2192. doi: 10.1093/genetics/165.4.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen DW, Gilbertson LG. Selective predation and intensity of selection acting upon lateral plates of threespine sticklebacks. Heredity. 1973;30:273–287. [Google Scholar]
- Haley CS, Knott SA. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity. 1992;69:315–324. doi: 10.1038/hdy.1992.131. [DOI] [PubMed] [Google Scholar]
- Harris MP, Rohner N, Schwarz H, Perathoner S, Konstantinidis P, Nusslein-Volhard C. Zebrafish eda and edar mutants reveal conserved and ancestral roles of ectodysplasin signaling in vertebrates. PLoS Genet. 2008;4(10):e1000206. doi: 10.1371/journal.pgen.1000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermisson J, Pennings PS. Soft Sweeps. Genetics. 2005;169:2335–2352. doi: 10.1534/genetics.104.036947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra HE, Coyne JA. The locus of evolution: evo devo and the genetics of adaptation. Evolution. 2007;61:995–1016. doi: 10.1111/j.1558-5646.2007.00105.x. [DOI] [PubMed] [Google Scholar]
- Hutchinson I, James TS, Clague JJ, Barrie JV, Conway KW. Reconstruction of late Quaternary sea-level change in southwestern British Columbia from sediments in isolation basins. Boreas. 2004;33:183–194. [Google Scholar]
- Ingram T, Svanback R, Kraft N, Kratina P, Southcott L, Schluter D. Intraguild predation drives evolutionary niche shift in threespine stickleback. Evolution. 2012 doi: 10.1111/j.1558-5646.2011.01545.x. In press. [DOI] [PubMed] [Google Scholar]
- Kassen R. Toward a general theory of adaptive radiation insights from microbial experimental evolution. Annals NY Acad. Sci. 2009;1168:3–22. doi: 10.1111/j.1749-6632.2009.04574.x. [DOI] [PubMed] [Google Scholar]
- Kaeuffer R, Peichel CL, Bolnick DI, Hendry AP. Parallel and nonparallel aspects of ecological, phenotypic, and genetic divergence across replicate population pairs of lake and stream stickleback. Evolution. 2012;66(2):305–616. doi: 10.1111/j.1558-5646.2011.01440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ullmann B, Walker C, Wilson C, Currey M, Phillips PC, Bell MA, Postlethwait JH, Cresko WA. Evolution and development of facial bone morphology in threespine sticklebacks. Proc. Natl. Acad. Sci. USA. 2005;102:5791–5796. doi: 10.1073/pnas.0408533102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp M, Hermisson J. Adaptation of a quantitative trait to a moving optimum. Genetics. 2007;176:715–719. doi: 10.1534/genetics.106.067215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langella O. [Accessed December 15, 2012];Populations: population genetic software (individuals or populations distances, phylogenetic trees) 2002 Available at http://bioinformatics.org/~tryphon/populations/
- Le Rouzic A, Ostbye K, Klepaker TO, Hansen TF, Bernatchez L, Schluter D, Vollestad LA. Strong and consistent natural selection associated with armour reduction in sticklebacks. Mol. Ecol. 2011;20:2483–2493. doi: 10.1111/j.1365-294X.2011.05071.x. [DOI] [PubMed] [Google Scholar]
- Leinonen T, Cano JM, Makinen H, Merila J. Contrasting patterns of body shape and neutral genetic divergence in marine and lake populations of threespine sticklebacks. J. Evol. Biol. 2006;19:1803–1812. doi: 10.1111/j.1420-9101.2006.01182.x. [DOI] [PubMed] [Google Scholar]
- Linnen CR, Hoekstra HE. Measuring natural selection on genotypes and phenotypes in the wild. Cold Spring Harb. Symp. Quant. Biol. 2010;74:155–168. doi: 10.1101/sqb.2009.74.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnen CR, Kingsley EP, Jensen JD, Hoekstra HE. On the origin and spread of an adaptive allele in deer mice. Science. 2009;325:1095–1098. doi: 10.1126/science.1175826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCairns RJS, Bernatchez L. Adaptive divergence between freshwater and marine sticklebacks: insights into the role of phenotypic plasticity from an integrated analysis of candidate gene expression. Evolution. 2010;64:1029–1047. doi: 10.1111/j.1558-5646.2009.00886.x. [DOI] [PubMed] [Google Scholar]
- McGregor AP, Orgogozo V, Delon I, Zanet J, Srinivasan DG, Payre F, Stern DL. Morphological evolution through multiple cis-regulatory mutations at a single gene. Nature. 2007;448:587–590. doi: 10.1038/nature05988. [DOI] [PubMed] [Google Scholar]
- Miller CT, Beleza S, Pollen AA, Schluter D, Kittles RA, Shriver MD, Kingsley DM. Cis-regulatory changes in kit ligand expression and parallel evolution of pigmentation in sticklebacks and humans. Cell. 2007;131:1179–1189. doi: 10.1016/j.cell.2007.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA. The population genetics of adaptation: the distribution of factors fixed during adaptive evolution. Evolution. 1998;52:935–949. doi: 10.1111/j.1558-5646.1998.tb01823.x. [DOI] [PubMed] [Google Scholar]
- Orr HA. The evolutionary genetics of adaptation: a simulation study. Genet. Res. 1999;74:207–214. doi: 10.1017/s0016672399004164. [DOI] [PubMed] [Google Scholar]
- Orr HA. The genetic theory of adaptation: a brief history. Nat. Rev. Genet. 2005;6:119–127. doi: 10.1038/nrg1523. [DOI] [PubMed] [Google Scholar]
- Orr HA, Coyne JA. The genetics of adaptation—a reassessment. Am. Nat. 1992;140:725–742. doi: 10.1086/285437. [DOI] [PubMed] [Google Scholar]
- Otto SP. Two steps forward, one step back: the pleiotropic effects of favoured alleles. Proc. R. Soc. Lond. Ser. B Biol. Sci. 2004;271:705–714. doi: 10.1098/rspb.2003.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peichel CL, Nereng KS, Ohgi KA, Cole BLE, Colosimo PF, Buerkle CA, Schluter D, Kingsley DM. The genetic architecture of divergence between threespine stickleback species. Nature. 2001;414:901–905. doi: 10.1038/414901a. [DOI] [PubMed] [Google Scholar]
- Peichel CL, Ross JA, Matson CK, Dickson M, Grimwood J, Schmutz J, Myers RM, Mori S, Schluter D, Kingsley DM. The master sex-determination locus in threespine sticklebacks is on a nascent Y chromosome. Curr. Biol. 2004;14:1416–1424. doi: 10.1016/j.cub.2004.08.030. [DOI] [PubMed] [Google Scholar]
- Phillips PC. Testing hypotheses regarding the genetics of adaptation. Genetica. 2005;123:15–24. doi: 10.1007/s10709-004-2704-1. [DOI] [PubMed] [Google Scholar]
- Pritchard JR, Schluter D. Declining interspecific competition during character displacement: summoning the ghost of competition past. Evol. Ecol. Res. 2001;3:209–220. [Google Scholar]
- R Development Team . R: a language and environment for statistical computing, reference index version R Foundation for Statistical Computing, Vienna, Austria. In: R. D. C. Team, editor. R Foundation for Statistical Computing. ISBN 3– 900051-07–0; Vienna , Austria: 2005. URL http://www.R-project.org. [Google Scholar]
- Renaut S, Nolte AW, Rogers SM, Derome N, Bernatchez L. SNP signatures of selection on standing genetic variation and their association with adaptive phenotypes along gradients of ecological speciation in lake whitefish species pairs (Coregonus spp.) Mol. Ecol. 2011;20:545–559. doi: 10.1111/j.1365-294X.2010.04952.x. [DOI] [PubMed] [Google Scholar]
- Rohlf FJ. tpsDig, digitize landmarks and outlines. Department of Ecology and Evolution, State University of New York at Stony Brook; 2005. Available at http://life.bio.sunysb.edu/ee/rohlf/software.html. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press; New York: 1989. [Google Scholar]
- Sawamura K, Davis A, Wu C. Genetic analysis of speciation by means of introgression into Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 2000;97:2652–2655. doi: 10.1073/pnas.050558597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter D, Conte GL. Genetics and ecological speciation. Proc. Natl. Acad. Sci. USA. 2009;106:9955–9962. doi: 10.1073/pnas.0901264106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter D, McPhail JD. Character displacement and replicate adaptive radiation. Trends Ecol. Evol. 1993;8:197–200. doi: 10.1016/0169-5347(93)90098-A. [DOI] [PubMed] [Google Scholar]
- Schluter D, Clifford EA, Nemethy M, McKinnon JS. Parallel evolution and inheritance of quantitative traits. Am. Nat. 2004;163:809–822. doi: 10.1086/383621. [DOI] [PubMed] [Google Scholar]
- Shapiro MD, Marks ME, Peichel CL, Blackman BK, Nereng KS, Jonsson B, Schluter D, Kingsley DM. Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature. 2004;428:717–723. doi: 10.1038/nature02415. [DOI] [PubMed] [Google Scholar]
- Stinchcombe JR, Hoekstra HE. Combining population genomics and quantitative genetics: finding the genes underlying ecologically important traits. Heredity. 2008;100:158–170. doi: 10.1038/sj.hdy.6800937. [DOI] [PubMed] [Google Scholar]
- Taylor E, McPhail J. Historical contingency and ecological determinism interact to prime speciation in sticklebacks, Gasterosteus. Proc. R. Soc. Lond. Ser. B Biol. Sci. 2000;267:2375–2384. doi: 10.1098/rspb.2000.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor EB, McPhail JD. Prolonged and burst swimming in anadromous and freshwater threespine stickleback, Gasterosteus aculeatus. Canadian Journal of Zoology. 1986;64:416–420. [Google Scholar]
- Vamosi SM. The presence of other fish species affects speciation in threespine sticklebacks. Evol. Ecol. Res. 2003;5:717–730. [Google Scholar]
- Van Ooijen JW, Voorrips RE. Plant Research International Wageningen. The Netherlands: 2001. JoinMap: software for the calculation of genetic linkage maps. [Google Scholar]
- Wahl LM, Krakauer DC. Models of experimental evolution: the role of genetic chance and selective necessity. Genetics. 2000;156:1437–1448. doi: 10.1093/genetics/156.3.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JA. Ecological morphology of lacustrine threespine stickleback Gasterosteus aculeatus L (Gasterosteidae) body shape. Biol. J. Linn. Soc. 1997;61:3–50. [Google Scholar]
- Walker JA, Bell MA. Net evolutionary trajectories of body shape evolution within a microgeographic radiation of threespine sticklebacks (Gasterosteus aculeatus) J. Zool. 2000;252:293–302. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.