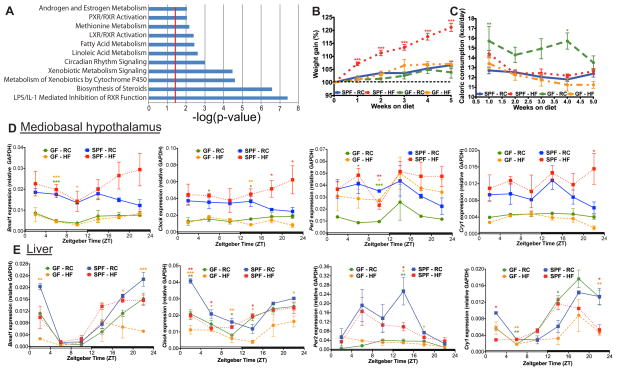
Figure 1. Germ-free mice exhibit differential canonical liver gene transcriptomic signatures and are resistant to diet-induced obesity.
GF and CONV C57Bl/6 liver transcriptomes generated via Affymetrix Mouse Genome 430 2.0 array (n=3 per group). Canonical pathways (A) enriched in GF versus CONV identified by Significant Analysis of Microarray software (FC >1.5 and FDR <5%) followed by submission to Ingenuity Pathway Analysis software. Significant pathways were determined by one-tail Fisher’s Exact test (p-value <0.05, i.e. −log(p-value) >1.3, (red line)). Weight gain (B) and food consumption (C) of SPF and GF mice (n=17–18 per group) fed RC or HF. Data represent mean ± s.e.m. ***p<0.001; **p<0.01; *p<0.05 via one-way ANOVA followed by Dunnett’s post-test relative to SPF-RC control where star color represents treatment exhibiting significance. Diurnal circadian gene expression patterns relative to GAPDH in mediobasal hypothalamus (D) and liver (E) in GF and SPF mice fed RC or HF. ***p<0.001; **p<0.01; *p<0.05 via one-way ANOVA followed by Dunnett’s post-test relative to SPF-RC control - star color indicates treatment exhibiting significance. See also Figure S1A–F and Table S2.