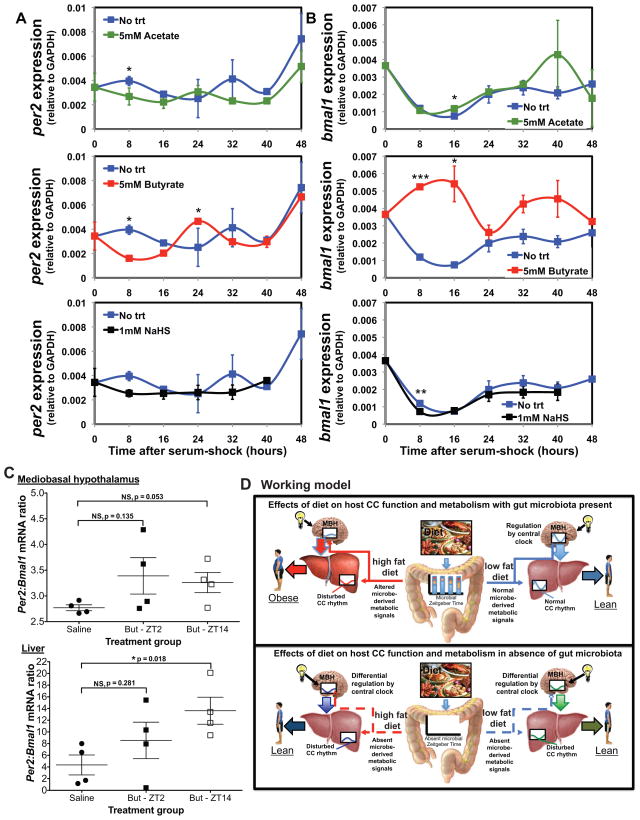
Figure 4. In vitro and in vivo exposure to diet-induced microbial metabolites alters host circadian gene expression.
Expression of per2 (A) and bmal1 (B) relative to GAPDH following addition of 5mM sodium acetate, 5mM sodium butyrate, or 1mM sodium hydrosulfide (NaHS) in hepanoids after serum-shock. Data is represented as mean ± s.e.m (n=3 replicates/treatment/time point). ***p<0.001; **p<0.01; and *p<0.05 determined via unpaired t-test compared to no trt within time point. See also Table S5. Ratio of per2:bmal1 mRNA (C) in MBH and liver of GF mice treated with saline or butyrate (But) at ZT2 or ZT14 for 5 days. Treatments are saline(ZT2)-saline(ZT14), saline(ZT2)-But(ZT14), But(ZT2)-saline(ZT14). Data represent mean ± s.e.m (n=4/treatment). *p<0.05; NS, not significant determined via unpaired t-test. Proposed experimental model (D) diet-induced change in gut microbe metabolic oscillatory patterns alters the balance between food consumption, the central CC, and hepatic regulatory networks of metabolism promoting DIO.