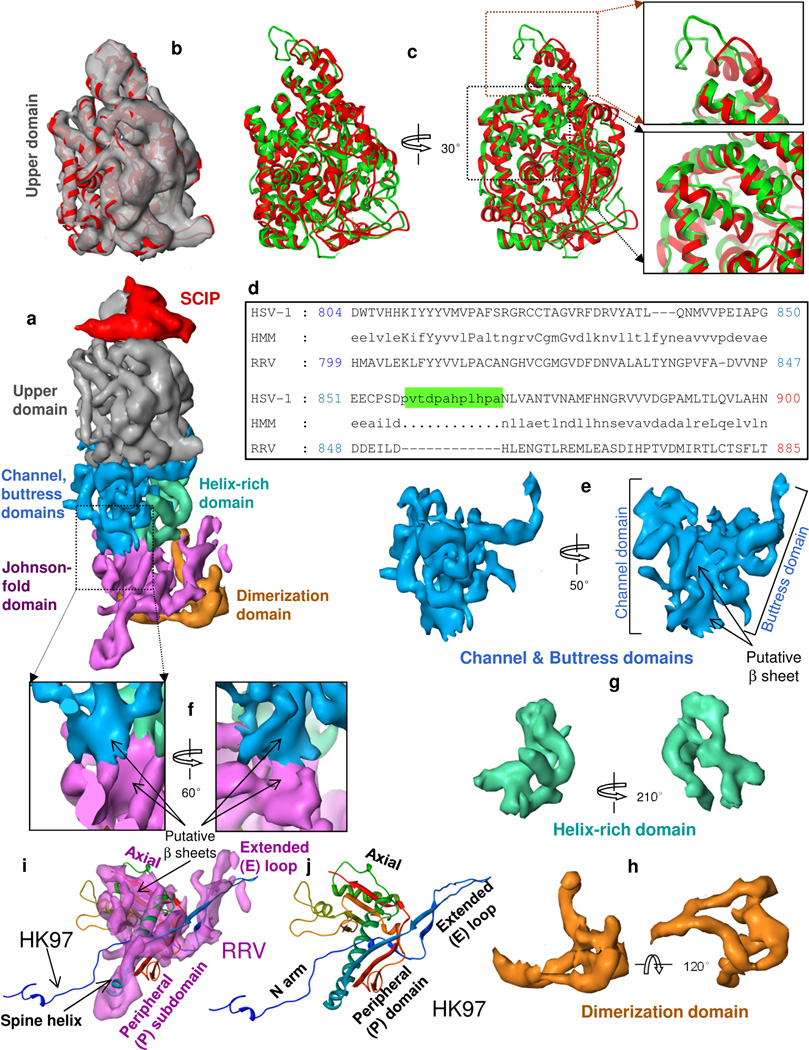
Figure 3. Domain organization of MCP.
(a) Side view of a hexon subunit. Each hexon subunit contains one monomer of SCIP (red) and one monomer of MCP. Each MCP monomer can be divided into six domains: upper domain in the upper region, channel, buttress and helix-rich domains in the middle region, and Johnson-fold and dimerization domains in the floor region. (b) Superposition of the cryoEM density map (semi-transparent gray) and a pseudo-atomic model (red ribbon) of the MCP upper domain (MCPud) of RRV. (c) The pseudo atomic model of the RRV MCPud (red) superimposed with the atomic model of HSV-1 MCPud (green). While most secondary structure elements in RRV and HSV-1 MCPud match (lower inset box), there are differences at the tip (upper inset box), possibly related to binding of SCIP and tegument proteins. (d) Alignment of the amino acid sequence between HSV-1 and RRV MCP in the tip of MCPud. HSV-1 MCP has a string of 12 additional amino acids (green highlighting). HMM: hidden Markov model, used to assist in alignment. (e–h) Surface representations of the channel and buttress domains (e), the helix-rich domain (g) and the dimerization domain (h). (i) Comparison of the surface representation of the Johnson-fold domain of RRV (semitransparent purple) with the atomic model of the Johnson fold of the bacteriophage HK97 (ribbon). (j) Atomic model of the HK97 gp5 protein (i.e., the Johnson fold), colored from N (blue) to C (red) termini. See also Fig. S2 and Movie S4.