Abstract
The genotypes and oxacillin resistance of 420 S. aureus isolates from pigs (n = 203) and pork (n = 217) were analyzed. Among 18 spa types detected in S. aureus from pig t011, t021, t034, t091, t318, t337, and t1334 were the most frequent. Among 30 spa types found in S. aureus isolates from pork t084, t091, t499, t4309, t12954, and t13074 were dominant. The animal S. aureus isolates were clustered into MLST clonal complexes CC7, CC9, CC15, CC30, and CC398 and meat-derived isolates to CC1, CC7, and CC15. Thirty-six MRSA were isolated exclusively from pigs. All MRSA were classified to spa t011 SCCmecV. BORSA phenotype was found in 14% S. aureus isolates from pigs and 10% isolates from pork meat. spa t034 dominated among BORSA from pigs and t091 among meat-derived BORSA. This is the first report on spa types and oxacillin resistance of S. aureus strains from pigs and pork meat in Poland. Besides S. aureus CC9, CC30, and CC398 known to be distributed in pigs, the occurrence of genotype belonging to CC7 in this species has been reported for the first time. To our knowledge it is also the first report concerning CC398 BORSA isolates from pigs and pork meat.
1. Introduction
Staphylococcus aureus is one of the most serious pathogens of humans and important animal pathogen. S. aureus infections can easily turn into life-threatening diseases if they are not antibiotically treated. The ability of this microorganism to survive in the presence of β-lactam antibiotics remains the main problem in the therapy [1]. Several phenotypes of resistance to β-lactams have been described in S. aureus so far. These phenotypes reflect different mechanisms of resistance and include acquisition of β-lactamase, modification of normal penicillin-binding proteins (PBPs), and acquisition of genes coding for low-drug-affinity PBPs. Methicillin-resistant S. aureus (MRSA), representing the latest of these aforementioned mechanisms, show resistance to both cefoxitin and oxacillin, which is conferred by acquisition of the mecA or recently discovered mecC gene [2, 3]. Another relatively frequently described phenotype amongst S. aureus strains is borderline oxacillin-resistant S. aureus (BORSA). These strains are cefoxitin susceptible and do not carry the mecA or mecC genes but are characterized by oxacillin resistance with MIC between 1 and 8 μg/mL [2, 3]. Hyperproduction of β-lactamase has been proposed to explain BORSA phenotype [4–6].
Increasing number of community-acquired MRSA (CA-MRSA) infections led to investigation of new sources of their origin. Current knowledge on population structure of cefoxitin-susceptible S. aureus from animals is still scarce. Some S. aureus genotypes are thought to be predominantly associated with particular animal species. The others can be isolated from both human and animals [7, 8]. Most of available data concern prevalence of animal MRSA [9–12]. Evidence for multiple, independent acquisition of the methicillin resistance determinant of methicillin-susceptible S. aureus (MSSA) strains, associated with animal breeding, enforces the research on revealing the structure of this population. Some of S. aureus lineages observed in animals are thought to arise from relatively recent transmission from humans. They may represent important reservoir of strains affecting global health systems. Research on genetic diversity of animal MSSA may allow identification of new clones potentially contributing to emergence of community-acquired staphylococcal infections [7].
Until recently MRSA were rarely isolated from livestock animals. However, in recent years, livestock-associated MRSA have been repeatedly isolated from pigs initially in Netherlands [13] and later in various countries in Europe [10], Canada [14], and USA [11]. Lineage ST398 was found to predominate among MRSA in pigs in Europe. It was observed that MRSA CC398 can be readily transferred from animal to animal and animal to human. Studies in humans showed that rapid transmission of MRSA CC398 is possible even after short-term occupational contact with colonized pigs [15].
spa genotyping scheme, first introduced by Harmsen et al. [16], enables interlaboratory comparison of genotypic data. Its association to the results of MLST typing allows for reliable genotypic characterization of S. aureus populations [17].
The aim of this work was to determine the population structure of S. aureus isolates derived from pig carriage and porcine meat, based on spa typing, with emphasis on detection of oxacillin-resistant isolates, that is, BORSA and MRSA.
2. Materials and Methods
2.1. Isolation and Identification of S. aureus
One thousand and seventy-four nasal swabs were taken between 2011 and 2012 from pigs in two slaughterhouses (S1 and S2) possessing their own meat processing plants located in south-west of Poland. Eight hundred and four nasal swabs were taken from the slaughterhouse S1 and 270 from the slaughterhouse S2. Average slaughter capacity was 200 pigs per day in both plants. The swabs were taken (ca. one hundred swabs per sampling) four times from slaughterhouse S1 and two times from S2 in 2011, as well as four times from S1 and once from S2 in 2012. Animals at each sampling session originated from different farms (11 objects in total). Samples were collected from the nasal cavity by introducing a cotton swab for approximately 10 cm into the nares. The swabs were taken after electric stunning, before steaming of the pigs. Slaughtered animals originated from local breeding farms.
Additionally, 396 and 140 samples of pork meat from S1 and S2 company shops were examined, respectively. Meat samples (ca. 20) were purchased from the company shops everyday successively during maximum of 4 days following the slaughter.
One-gram food samples and whole nasal cotton swabs were cultured in a final volume of 10 mL of Giolitti-Cantoni enrichment broth and subcultured on Baird-Parker agar. The isolates were identified as S. aureus based on their ability to coagulate rabbit plasma and clumping factor production. All isolates were screened by PCR using S. aureus-specific primers for nuc gene, encoding thermonuclease [18]. Reference S. aureus strain ATCC 29213 served as a control. One S. aureus isolate per sample/swab was taken for further characterization.
2.2. Preparation of Bacterial DNA
Two millilitres of bacterial cell suspension from an overnight culture grown in brain-heart infusion (BHI) broth was centrifuged for 5 min at 12,000 ×g and suspended in 100 μL of 100 mM Tris-HCl buffer, pH 7.4, containing 10 μg of lysostaphin (A&A Biotechnology, Gdańsk, Poland). After 30-minute incubation at 37°C, 10 μL of 10% SDS was added and the sample was incubated for another 30 min at 37°C. Two hundred μL of 5 M guanidine hydrochloride was added and the sample was mixed by vortexing and incubated at room temperature for 10 min. The DNA was extracted by phenol and chloroform, precipitated with ethanol, and dissolved in water.
2.3. Detection of mecA and Determination of SCCmec Cassette Type
All S. aureus isolates were tested for the presence of mecA gene using the primers described by Milheiriço et al. [19]. Each PCR contained mecA-positive (S. aureus ATCC 43300) and -negative (S. aureus ATCC 29213) strains as controls. SCCmec cassette type was determined according to Milheiriço et al. [19]. The PCR products were electrophoretically resolved in 1.5% agarose containing 0.5 μg/mL ethidium bromide and photographed with the GelDocXR System (Bio-Rad, Hercules, CA).
2.4. Antibiotic Resistance and Oxacillin MIC Determination in mecA-Positive S. aureus
Susceptibility of mecA-positive S. aureus isolates to penicillin G (10 units/disc), cefoxitin (30 μg/disc), tetracycline (30 μg/disc), clindamycin (2 μg/disc), gentamicin (10 μg/disc), erythromycin (15 μg/disc), ciprofloxacin (5 μg/disc), norfloxacin (10 μg/disc), and vancomycin (30 μg/disc) (all substances from Oxoid Ltd., UK) was tested by the disk-diffusion method and interpreted according to CLSI document M100-S22 [20]. The MIC for oxacillin was determined with the E-test and interpreted according to the manufacturer's instructions (bio-Mérieux, Inc.). Reference S. aureus strains ATCC 25923, ATCC 43300, and ATCC 29213 served as controls.
2.5. Detection of Borderline Oxacillin-Resistant S. aureus (BORSA)
All mecA-negative S. aureus isolates were plated on oxacillin resistance screening agar (ORSA, Oxoid) plates containing 2, 3, and 4 μg/mL oxacillin, respectively. The results were recorded after 24- and 48-hour incubation at 35°C and interpreted according to the manufacturer's instructions. For isolates able to grow in medium containing 4 μg/mL oxacillin MIC for oxacillin was determined using E-test (bio-Mérieux, Inc.). Reference MRSA (ATCC 43300) and MSSA (ATCC 29213) strains served as controls. All phenotypically oxacillin-resistant isolates were analyzed for susceptibility to amoxicillin with clavulanic acid (20/10 μg/disc) by the disk-diffusion method and interpreted according to CLSI document M100-S22 [20]. Reference E. coli ATCC 35218 strain served as control. All BORSA isolates were screened for blaZ gene according to Rizzotti et al. [21] and for hyperproduction of beta-lactamase using Cefinase test (bio-Mérieux, Inc.).
2.6. Detection of mecC
All mecA-negative S. aureus isolates able to grow on ORSA plates containing 2 μg/mL oxacillin were tested for mecC gene using the primers described by Cuny et al. [22]. DNA from mecC-positive S. aureus strain 1140/12, from the National Medicines Institute, Warsaw, Poland, served as a control.
2.7. Determination of spa Type and ST
The spa types of all S. aureus isolates were determined according to Harmsen et al. [16]. The nucleotide sequencing of the repeat-containing region of the spa gene was performed from both DNA strands of the PCR product by Genomed (Warsaw, Poland), using BigDye Terminator Ready Reaction Cycle Sequencing kit. The analysis of repeats and the assignment of spa types were performed with the resources of the Ridom SpaServer (http://spa.ridom.de). Grouping of spa types was done using BURP, Ridom Staphtype Software. Spa types were clustered if cost between members of the group was less than or equal to 4. Spa types shorter than 5 repeats were excluded from analysis [23].
Sequence types (STs) of selected S. aureus isolates (one t091 isolate from pig and 9 isolates from meat, i.e., t091, t118, t289, t519, t3358, t9031, t12953, t12954, and t12955) were determined according to Enright et al. [24]. The sequences obtained from both strands of the PCR product were analyzed using BioEdit software (http://www.mbio.ncsu.edu/bioedit/bioedit.html) and further assignment of the sequence type (ST) was performed using the http://www.mlst.net/ platform.
3. Results
3.1. Frequency of S. aureus Isolates
In total, 420 S. aureus isolates were obtained, including 203 isolates from 1074 nasal swabs and 217 isolates from 536 meat samples. The prevalence of S. aureus was different in the two slaughterhouses. The bacterium was found in 197 (25%) from a total of 804 nasal swabs in S1, but only in 6 (2%) from 270 swabs in S2. Screening of meat from S1 and S2 company shops revealed comparable prevalence of samples contaminated with S. aureus. The pathogen was isolated from 157 (40%) of a total of 396 meat samples derived from S1 and 60 (43%) from 140 samples originating from S2.
3.2. Genotypes of S. aureus Isolates
Forty-three spa types were determined in studied S. aureus population. S. aureus isolates obtained from pig nasal swabs were classified into 18 spa types. Among them t318 (28.7%), t011 (18.3%), t034 (13.4%), t337 (11.9%), t021 (8.9%), t091 (8.4%), and t1334 (3.0%) were most frequent (Table 3), with genotypes t318, t034, and t091 isolated during 5 out of a total of 11 sampling sessions (Tables 1 and 2).
Table 3.
Clonal spa complexes of S. aureus isolates from pig nasal swabs and pork meat.
| Cluster | spa | spa-CC | MLST-CC | n | Origin |
|---|---|---|---|---|---|
| 1 | t091 | spa-CC 499 | CC7* | 108 | Nasal swab (n = 17) and pork meat (n = 91) |
| 1 | t4309 | spa-CC 499 | CC15 | 32 | Nasal swab (n = 1) and pork meat (n = 31) |
| 1 | t084 | spa-CC 499 | CC15 | 26 | Nasal swab (n = 1) and pork meat (n = 25) |
| 1 | t499 | spa-CC 499 | CC15 | 12 | Pork meat |
| 1 | t346 | spa-CC 499 | CC15 | 1 | Pork meat |
| 1 | t289 | spa-CC 499 | CC7* | 1 | Pork meat |
| 1 | t774 | spa-CC 499 | CC15 | 1 | Pork meat |
| 1 | t7568 | spa-CC 499 | CC15 | 1 | Nasal swab |
| 2 | t337 | spa-CC 1334 | CC9 | 26 | Nasal swab (n = 24) and pork meat (n = 2) |
| 2 | t1334 | spa-CC 1334 | CC9 | 7 | Nasal swab |
| 2 | t1430 | spa-CC 1334 | CC9 | 4 | Nasal swab |
| 2 | t12950 | spa-CC 1334 | CC9 | 2 | Nasal swab |
| 2 | t8893 | spa-CC 1334 | CC9 | 1 | Nasal swab |
| 2 | t12952 | spa-CC 1334 | CC9 | 1 | Nasal swab |
| 3 | t13074 | spa-CC 273 | CC1 | 8 | Pork meat |
| 3 | t1491 | spa-CC 273 | CC1 | 6 | Pork meat |
| 3 | t273 | spa-CC 273 | CC1 | 3 | Pork meat |
| 4 | t034 | spa-CC 034 | CC398 | 31 | Nasal swab (n = 27) and pork meat (n = 4) |
| 4 | t011 | spa-CC 034 | CC398 | 37 | Nasal swab |
| 4 | t8588 | spa-CC 034 | CC398 | 1 | Nasal swab |
| 5 | t12954 | spa-CC 12954 | CC1* | 12 | Pork meat |
| 5 | t12955 | spa-CC 12954 | CC1* | 1 | Pork meat |
| 5 | t12953 | spa-CC 12954 | CC1* | 1 | Pork meat |
| 6 | t1333 | spa-CC 021 | CC30 | 1 | Pork meat |
| 6 | t318 | spa-CC 021 | CC30 | 58 | Nasal swab |
| 6 | t021 | spa-CC 021 | CC30 | 18 | Nasal swab |
| 7 | t304 | spa-CC 008 | CC8 | 1 | Pork meat |
| 7 | t1187 | spa-CC 008 | CC8 | 1 | Pork meat |
| 7 | t008 | spa-CC 008 | CC8 | 1 | Pork meat |
| Singleton | t3478 | CC5 | 2 | Pork meat | |
| Singleton | t3380 | CC1 | 2 | Pork meat | |
| Singleton | t3358 | CC101* | 2 | Pork meat | |
| Singleton | t015 | CC45 | 1 | Pork meat | |
| Singleton | t078 | CC25 | 1 | Pork meat | |
| Singleton | t127 | CC1 | 1 | Pork meat | |
| Singleton | t156 | CC12 | 1 | Pork meat | |
| Singleton | t267 | CC97 | 1 | Pork meat | |
| Singleton | t9031 | ST1027* | 1 | Pork meat | |
| Singleton | t252 | CC15 | 1 | Nasal swab | |
| Excluded | t118 | ST2811* | 1 | Pork meat | |
| Excluded | t519 | CC7* | 1 | Pork meat | |
| Excluded | t1939 | CC398 | 1 | Nasal swab | |
| Excluded | t026 | CC45 | 1 | Nasal swab | |
|
| |||||
| Total spa | 43 | Total isolates | 420 | ||
MLST-CCs marked with asterisk (∗) were determined in this study as described by Enright et al. (2000) [24]. For the remaining MLST-CCs, associations with the particular spa types were assessed through the Ridom SpaServer (http://spa.ridom.de) or from the relevant literature.
Table 1.
spa types of S. aureus isolates from pig nasal swabs and pork meat derived from slaughterhouses S1 and S2 in 2011.
| Sampling | Nasal swabs | Pork meat | ||||
|---|---|---|---|---|---|---|
| Number of samples | Number of isolates | spa types (n) | Number of samples | Number of isolates | spa types (n) | |
| S. aureus isolates from S1 | ||||||
| Sampling I | 101 | 44 | t318 (29), t034 (7), t1430 (3), t091 (2), t7568 (1), t4309 (1), t8588 (1) | 95 | 32 | t091 (12), t084 (12), t4309 (3), t273 (1), t499 (2), t015 (1), t118 (1) |
| Sampling II | 101 | 19 | t021 (15), t318 (3), t034 (1) | 65 | 11 | t091 (7), t084 (2), t273 (2) |
| Sampling III | 100 | 0 | — | Not tested | 0 | — |
| Sampling IV | 100 | 2 | t1334 (2) | 36 | 0 | — |
| Total | 402 | 65 | 196 | 43 | ||
|
| ||||||
| S. aureus isolates from S2 | ||||||
| Sampling I | 100 | 1 | t252 (1) | 50 | 21 | t091 (6), t084 (4), t034 (2), t499 (2), t156 (1), t289 (1), t304 (1), t346 (1), t519 (1), t1491 (1), t3358 (1) |
| Sampling II | 100 | 3 | t12950 (2), t1430 (1) | 50 | 22 | t084 (6), t1491 (5), t091 (4), t3478 (2), t034 (1), t127 (1), t267 (1), t337 (1), t499 (1) |
| Total | 200 | 4 | 100 | 43 | ||
Table 2.
spa types of S. aureus isolates from pig nasal swabs and pork meat derived from slaughterhouses S1 and S2 in 2012.
| Sampling | Nasal swabs | Pork meat | ||||
|---|---|---|---|---|---|---|
| Number of samples | Number of isolates | spa types (n) | Number of samples | Number of isolates | spa types (n) | |
| S. aureus isolates from S1 | ||||||
| Sampling I | 101 | 39 | t318 (24), t034 (6), t091 (4), t1334 (4), t1939 (1) | 50 | 26 | t4309 (11), t091 (10), t13074 (2), t034 (1), t1187 (1), t9031 (1) |
| Sampling II | 101 | 49 | t011 (37), t091 (4), t021 (3), t034 (2), t026 (1), t084 (1), t1334 (1) | 50 | 31 | t091 (9), t4309 (7), t12954 (7), t499 (3), t084 (1), t337 (1), t12955 (1), t13074 (1) |
| Sampling III | 100 | 32 | t337 (21), t034 (9), t091 (1), t318 (1) | 50 | 27 | t091 (15), t4309 (7), t13074 (2), t078 (1), t499 (1), t12954 (1) |
| Sampling IV | 100 | 12 | t091 (6), t337 (3), t034 (2), t8893 (1) | 50 | 30 | t091 (19), t12954 (4), t4309 (3), t13074 (2), t499 (1), t774 (1) |
| Total | 402 | 132 | 200 | 114 | ||
|
| ||||||
| S. aureus isolates from S2 | ||||||
| Sampling I | 70 | 2 | t318 (1), t12952 (1) | 40 | 17 | t091 (9), t499 (2), t3380 (2), t008 (1), t1333 (1), t3358 (1), t12953 (1) |
| Total | 70 | 2 | 40 | 17 | ||
S. aureus isolates from pork meat were assigned to 30 spa types. Among them isolates belonging to t091 (41.7%), t4309 (14.2%), t084 (11.5%), t499 (5.5%), t12954 (5.5%), and t13074 (3.7%) were dominating (Table 3). S. aureus genotype t091 was isolated during 9 out of 11 sampling sessions, while t4309 was found in samples from 5 sessions (Tables 1 and 2).
The most abundant genotype amongst isolates from animals, t318 and t011, were not found in food (Tables 1 and 2). spa genotypes t034 and t337 were frequently isolated from animals (13.3% and 11.8%, resp.), but sporadically from meat (1.8% and 0.9%, resp.). In contrast, genotypes t4309 and t084 occurred sporadically in pigs (0.5% each) but were frequent in pork meat (14.3% and 11.5%, resp.) (Tables 1 and 2). Thirteen out of 18 S. aureus genotypes identified in animals were not detected in meat, whereas among 30 spa types found in meat 25 were not detected in animals. Only 5 S. aureus genotypes, that is, t034, t084, t091, t337, and t4309, were identified in both nasal swabs and meat (Table 3).
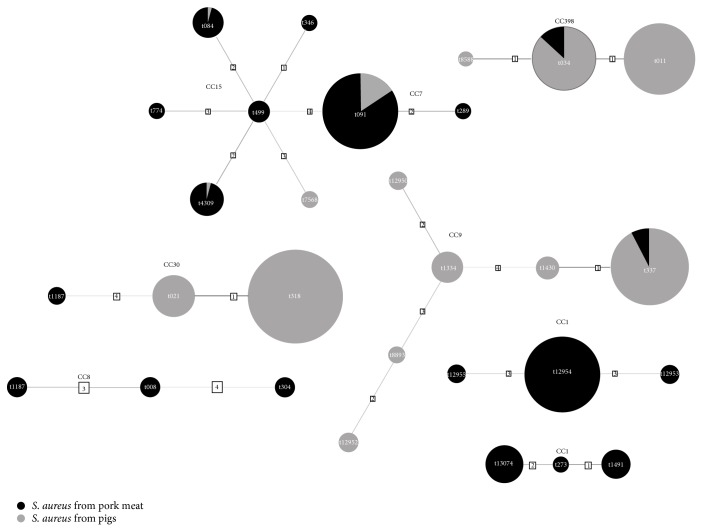
All genotypes identified in the studied S. aureus isolates were clustered into 7 spa complexes (Figure 1). The animal isolates were clustered into four complexes, namely, spa-CC034 (t011, t034, and t8588) belonging to ST-CC398, spa-CC1334 (t337, t1334, t1430, t8893, t12950, and t12952) within ST-CC9, spa-CC021 (t021, t318) within ST-CC30, and spa-CC499 (t084, t091, t4309, and t7568) within ST-CC7 and ST-CC15 (Figure 1). The spa genotypes of meat-derived S. aureus isolates were clustered into 7 complexes. Most isolates (73.4%) were grouped into spa-CC499 belonging to ST-CC15 and ST-CC7. Other numerous complexes included spa-CC273 (7.8%) and newly described spa-CC12954 (6.4%) both belonging to ST-CC1 (Figure 1, Table 3).
Figure 1.
Cluster analysis of spa types of S. aureus isolates from pig nasal swabs and pork meat.
3.3. Characterization of MRSA Isolates
Thirty-six (17.8%) S. aureus isolates from pigs were classified as MRSA. All these isolates were mecA-positive and resistant to cefoxitin in the disc-diffusion method. None of them possessed mecC gene. All of the isolates were resistant to oxacillin with MIC ranged from 32 to 48 μg/mL. Additionally all of the MRSA isolates were resistant to penicillin and tetracycline and susceptible to gentamicin, erythromycin, ciprofloxacin, norfloxacin, and vancomycin. MRSA were isolated exclusively from pigs in slaughterhouse S1, originating from a single farm. All of them were classified to spa type t011 SCCmecV.
3.4. Characterization of BORSA Isolates
Twenty-eight (14%) S. aureus isolates from pigs and 21 (10%) from meat exhibited borderline resistance to oxacillin. All of them were mecA and mecC negative, susceptible to cefoxitin and amoxicillin with clavulanic acid, and able to grow on ORSA plates containing minimum of 2 μg/mL oxacillin. Seventeen (30%) and 3 (5%) of BORSA isolates grew on 3 and 4 μg/mL oxacillin, respectively. MIC for oxacillin was <5 μg/mL in isolates able to grow on 4 μg/mL oxacillin. All BORSA isolates were positive in Cefinase test and harboured blaZ gene. spa t034 was found to dominate among BORSA from pigs (64%) and t091 among meat-derived BORSA (38%) (Table 4).
Table 4.
Characteristics of BORSA isolates from pig nasal swabs and pork meat.
| Oxacillin concentration permitting bacterial growth | Nasal swabs | Pork meat | ||
|---|---|---|---|---|
| spa type | Number of isolates | spa type | Number of isolates | |
| 4 μg/mL | t034 | 1 | t091 | 2 |
|
| ||||
| 3 μg/mL | t034 | 6 | t091 | 6 |
| t011 | 1 | t034 | 1 | |
| t8588 | 1 | t13074 | 1 | |
| t337 | 1 | |||
|
| ||||
| 2 μg/mL | t034 | 11 | t12954 | 4 |
| t318 | 1 | t273 | 2 | |
| t337 | 4 | t1491 | 1 | |
| t026 | 1 | t034 | 1 | |
| t1334 | 1 | t127 | 1 | |
| t1187 | 1 | |||
| t13074 | 1 | |||
|
| ||||
| Total 28 | Total 21 | |||
4. Discussion
Animal production models, that is, concentration of production in limited number of big holdings or in numerous small farms, are thought to influence the structure of S. aureus population [7]. Large European screening of pooled dust samples from pig breeding farms which focused on MRSA demonstrated considerable variation in terms of MRSA prevalence and their genotypes among EU countries [10]. Our previous research indicated low incidence of oxacillin-resistant S. aureus in food of animal origin in Poland [25]. This encouraged us to investigate the incidence and genotypes of S. aureus in pigs and pork meat in south-western Poland.
Our data indicate 19% mean incidence of S. aureus in pigs and 40% in pork meat. However, it should be stressed that depending on sampling session it varied from 0% to 48% in animals and from 0 to 44% in meat indicating significant variation in S. aureus occurrence among farms. As yet most research on incidence of S. aureus in pigs was focussed on MRSA [7]. Studies, like that by Vandendriessche et al. [26], carried out in Belgium, demonstrating MSSA occurrence (27%) and spa genotype structure among pigs are still rare.
S. aureus genotypes from pigs studied here clustered into four clonal complexes, namely, CC30 (38% of all animal isolates), CC398 (32%), CC9 (19%), and CC7 (8%). High prevalence of MSSA from genetic lineages CC30 and CC398 has been already reported in pigs in Europe [26, 27]. Animal contamination with ST398 MRSA increased over last years. A number of evidences demonstrate the possibility of their transmission to humans [28]. Data from EFSA report indicate t011 MRSA as most frequent in Europe [10]. Similarly our MRSA isolates of spa type t011 constituted most numerous populations within CC398, but it should be emphasized that they were found at one sampling only. According to the only data on CC398 MRSA associated with pig environment in Poland their prevalence was not higher than 2% [10]. Here we demonstrate that although overall prevalence of genotype t011 MRSA in pigs was ca. 3%, their occurrence was likely restricted to single source. In turn, t034 MSSA was the most prevalent genotype (32%) belonging to CC398 in Denmark [27]. Our t034 MSSA isolates, although less numerous (12%), consistently occurred in a series of sessions, what may indicate wide dissemination of this genotype in Poland.
Transmission of CC398 from animals to food was not extensively investigated as yet. CC398 MRSA was already detected in milk and meat [28, 29]. According to some surveys pork meat contamination with CC398 strains was relatively frequent. Results of a Dutch survey report MRSA t011 isolates in 7% of pork meat [28]. Spanish report indicates 3% frequency of ST398 MRSA in raw pork [30]. We could not detect MRSA in pork meat; however our previous surveys on MRSA incidence in food of animal origin support very low frequency of these strains in Poland [25, 31]. Results presented here demonstrated, however, that MSSA and BORSA t034 isolates could be found in pork meat indicating potential of sporadic introduction of animal-associated genotypes into food chain.
Although the incidence of BORSA among human clinical isolates was reported to be about 5%, they have been implicated in community-acquired and hospital infections [32–34]. BORSA have already been detected in food primarily associated with ruminant milk [25, 35]. In turn, cows seem to be the only animal species in which incidence of BORSA was reported [36]. Genotypic structure of human BORSA population is largely unknown, and there are no data on animal BORSA genotypes. In the current study, 28 and 21 BORSA isolates from pigs and pork meat were identified, respectively. As much as 71% of animal BORSA isolates and 10% of meat isolates were assigned to CC398. These results illustrate the possibility of transmission of typical animal-associated, oxacillin-resistant S. aureus isolates to food. Remaining numerous BORSA genotypes belong to CC9 in animal isolates, as well as to CC1 and CC7 in meat isolates.
Another major spa cluster identified in this study within animal S. aureus isolates, including t1334, t337, t1430, t8893, t12950, and 12952, belongs to CC9. LA-MRSA and MSSA of CC9 are frequent in livestock in Asia [12, 37, 38]. Some reports confirm their low-rate occurrence in Europe [26, 27, 39]. As yet the only European animal spa types identified within ST9 include t1430, found in poultry in Netherlands [40], t337 isolated from pigs in Denmark and Belgium [26, 27], and t4794 MRSA from Italy [39]. In turn, spa types t337 and t899, representing CC9, were the main pig-derived MRSA in Thailand [38] and Hong Kong [12].
Incidence of t091 isolates, representing genotype belonging to ST7 (CC7), which consequently occurred in studied here pigs at several sampling sessions has not been already reported in this species, suggesting the possibility of emergence of new genotype in livestock. t091 CA-MSSA together with t084 and t774 belonging to CC15, also found in studied animals, have been isolated only from human as yet [41] and recently found in broilers [26]. On the other side, isolates of CC7 and CC15 predominated in pork meat investigated here, accounting for 73% of total isolates derived from this product, suggesting additional most likely human source of meat contamination.
Isolates belonging to CC1 (almost 14%), including isolates assigned to new spaCC 12954 type, and CC8 (1.3%) were detected exclusively in meat. Although sporadic occurrence of these genotypes in livestock was already noted [10, 42, 43] isolation of these clones has been reported mainly from human [41, 44].
Only five genotypes, that is, t091, t034, t337, t084, and t4309, were isolated from both animals and pork meat. From these only t091 was identified with high frequency in both populations, while other genotypes consistently predominated in pigs (t034, t337) or meat (t084, t4309). Taking into account a significant diversity of bacterial genotypes revealed in consecutive samplings we cannot draw definite conclusions on the adaptation of studied genotypes to animal or food milieu. However, results of this survey generally demonstrating a weak genotypic relatedness of S. aureus from pigs and isolates from pork meat may indicate that certain genotypes can be introduced more readily than others into food chain. It seems that t091 genotype isolates can be better fitted to colonize both pigs and porcine food products. Although our data showed additional nonanimal source of meat contamination, as discussed above for CC1, CC7, CC8, and CC15 isolates, meat contamination may also be associated with S. aureus derived from niches other than pig nares, like skin or intestinal tract. Additional research should be conducted to shed light on structure of S. aureus carriage in animals.
5. Conclusions
Taking together, we first report on spa types and the oxacillin resistance of S. aureus isolates from pigs and pork meat in Poland. Besides S. aureus CC30, CC398, and CC9 already known to be widely distributed in pigs, genotype t091 belonging to CC7 was first reported to occur in this species. This is also the first report on the occurrence of CC398 BORSA isolates in pigs and MSSA and BORSA CC398 isolates in pork meat. CC7 isolates, including BORSA phenotype, together with isolates assigned to CC15 were shown to dominate in pork meat.
Acknowledgment
This work was financially supported by the National Science Centre, Poland, Decision no. 2581/B/P01/2011/40.
Conflict of Interests
None of the authors of this paper has a financial or personal relationship with other people or organizations that could inappropriately influence or bias the content of the paper.
References
- 1.Pinho M. G. Staphylococcus Molecular Genetics. Norfolk, Va, USA: Caister Academic Press; 2008. [Google Scholar]
- 2.Chambers H. F. Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. Clinical Microbiology Reviews. 1997;10(4):781–791. doi: 10.1128/cmr.10.4.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shore A. C., Coleman D. C. Staphylococcal cassette chromosome mec: recent advances and new insights. International Journal of Medical Microbiology. 2013;303(6-7):350–359. doi: 10.1016/j.ijmm.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 4.McDougal L. K., Thornsberry C. The role of β-lactamase in staphylococcal resistance to penicillinase-resistant penicillins and cephalosporins. Journal of Clinical Microbiology. 1986;23(5):832–839. doi: 10.1128/jcm.23.5.832-839.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tomasz A., Drugeon H. B., de Lencastre H. M., Jabes D., McDougall L., Bille J. New mechanism for methicillin resistance in Staphylococcus aureus: clinical isolates that lack the PBP2a gene and contain normal penicillin-binding proteins with modified penicillin-binding capacity. Antimicrobial Agents and Chemotherapy. 1989;33(11):1869–1874. doi: 10.1128/aac.33.11.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michel M., Gutmann L. Methicillin-resistant Staphylococus aureus and vancomycin-resistant enterococci: therapeutic realities and possibilities. The Lancet. 1997;349(9069):1901–1906. doi: 10.1016/s0140-6736(96)11192-2. [DOI] [PubMed] [Google Scholar]
- 7.Fitzgerald J. R. Livestock-associated Staphylococcus aureus: origin, evolution and public health threat. Trends in Microbiology. 2012;20(4):192–198. doi: 10.1016/j.tim.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 8.Mccarthy A. J., Lindsay J. A., Loeffler A. Are all meticillin-resistant Staphylococcus aureus (MRSA) equal in all hosts? Epidemiological and genetic comparison between animal and human MRSA. Veterinary Dermatology. 2012;23(4):267–e54. doi: 10.1111/j.1365-3164.2012.01072.x. [DOI] [PubMed] [Google Scholar]
- 9.de Neeling A. J., van den Broek M. J. M., Spalburg E. C., et al. High prevalence of methicillin resistant Staphylococcus aureus in pigs. Veterinary Microbiology. 2007;122(3-4):366–372. doi: 10.1016/j.vetmic.2007.01.027. [DOI] [PubMed] [Google Scholar]
- 10.EFSA. Analysis of the baseline survey on the prevalence of methicillin-resistant Staphylococcus aureus (MRSA) in holdings with breeding pigs, in the EU, 2008. Part A: MRSA prevalence estimates. EFSA Journal. 2009;7(11, article 1376) [Google Scholar]
- 11.Smith T. C., Male M. J., Harper A. L., et al. Methicillin-resistant Staphylococcus aureus (MRSA) strain ST398 is present in midwestern U.S. swine and swine workers. PLoS ONE. 2009;4(1) doi: 10.1371/journal.pone.0004258.e4258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ho J., O'Donoghue M., Guardabassi L., Moodley A., Boost M. Characterization of methicillin-resistant Staphylococcus aureus isolates from pig carcasses in Hong Kong. Zoonoses and Public Health. 2012;59(6):416–423. doi: 10.1111/j.1863-2378.2012.01473.x. [DOI] [PubMed] [Google Scholar]
- 13.Voss A., Loeffen F., Bakker J., Klaassen C., Wulf M. Methicillin-resistant Staphylococcus aureus in Pig Farming. Emerging Infectious Diseases. 2005;11(12):1965–1966. doi: 10.3201/eid1112.050428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khanna T., Friendship R., Dewey C., Weese J. S. Methicillin resistant Staphylococcus aureus colonization in pigs and pig farmers. Veterinary Microbiology. 2008;128(3-4):298–303. doi: 10.1016/j.vetmic.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 15.Van Cleef B. A. G. L., Graveland H., Haenen A. P. J., et al. Persistence of livestock-associated methicillin-resistant Staphylococcus aureus in field workers after short-term occupational exposure to pigs and veal calves. Journal of Clinical Microbiology. 2011;49(3):1030–1033. doi: 10.1128/jcm.00493-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harmsen D., Claus H., Witte W., Rothgänger J., Turnwald D., Vogel U. Turnwald and U. Vogel, 2003: typing of methicillin-resistant Staphylococcus aureus in a university hospital setting using a novel software for spa-repeat determination and database management. Journal of Clinical Microbiology. 2003;41(12):5442–5448. doi: 10.1128/jcm.41.12.5442-5448.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strommenger B., Kettlitz C., Weniger T., Harmsen D., Friedrich A. W., Witte W. Assignment of Staphylococcus isolates to groups by spa typing, SmaI macrorestriction analysis, and multilocus sequence typing. Journal of Clinical Microbiology. 2006;44(7):2533–2540. doi: 10.1128/jcm.00420-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martín M. C., González-Hevia M. A., Mendoza M. C. Usefulness of a two-step PCR procedure for detection and identification of enterotoxigenic staphylococci of bacterial isolates and food samples. Food Microbiology. 2003;20(5):605–610. doi: 10.1016/S0740-0020(02)00180-6. [DOI] [Google Scholar]
- 19.Milheiriço C., Oliveira D. C., de Lencastre H. Update to the multiplex PCR strategy for assignment of mec element types in Staphylococcus aureus . Antimicrobial Agents and Chemotherapy. 2007;51(9):3374–3377. doi: 10.1128/aac.00275-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.CLSI. Document. M100-S22. Wayne, Pa, USA: Clinical and Laboratory Standards Institute; 2012. Performance standards for antimicrobial susceptibility testing; twenty-second informational supplement. [Google Scholar]
- 21.Rizzotti L., Simeoni D., Cocconcelli P., Gazzola S., Dellaglio F., Torriani S. Contribution of enterococci to the spread of antibiotic resistance in the production chain of swine meat commodities. Journal of Food Protection. 2005;68(5):955–965. doi: 10.4315/0362-028x-68.5.955. [DOI] [PubMed] [Google Scholar]
- 22.Cuny C., Layer F., Strommenger B., Witte W. Rare occurrence of methicillin-resistant Staphylococcus aureus CC130 with a novel mecA homologue in humans in Germany. PLoS ONE. 2011;6(9) doi: 10.1371/journal.pone.0024360.e24360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mellmann A., Weniger T., Berssenbrügge C., et al. Based Upon Repeat Pattern (BURP): an algorithm to characterize the long-term evolution of Staphylococcus aureus populations based on spa polymorphisms. BMC Microbiology. 2007;7, article 98 doi: 10.1186/1471-2180-7-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Enright M. C., Day N. P. J., Davies C. E., Peacock S. J., Spratt B. G. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of (MSSA) Staphylococcus aureus . Journal of Clinical Microbiology. 2000;38(3):1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bystroń J., Podkowik M., Korzekwa K., Lis E., Molenda J., Bania J. Characterization of borderline oxacillin-resistant Staphylococcus aureus isolated from food of animal origin. Journal of Food Protection. 2010;73(7):1325–1327. doi: 10.4315/0362-028x-73.7.1325. [DOI] [PubMed] [Google Scholar]
- 26.Vandendriessche S., Vanderhaeghen W., Larsen J., et al. High genetic diversity of methicillin-susceptible Staphylococcus aureus (MSSA) from humans and animals on livestock farms and presence of SCCmec remnant DNA in MSSA CC398. Journal of Antimicrobial Chemotherapy. 2014;69(2):355–362. doi: 10.1093/jac/dkt366. [DOI] [PubMed] [Google Scholar]
- 27.Hasman H., Moodley A., Guardabassi L., Stegger M., Skov R. L., Aarestrup F. M. spa type distribution in Staphylococcus aureusoriginating from pigs, cattle and poultry. Veterinary Microbiology. 2010;141(3-4):326–331. doi: 10.1016/j.vetmic.2009.09.025. [DOI] [PubMed] [Google Scholar]
- 28.Verkade E., Kluytmans J. Livestock-associated Staphylococcus aureus CC398: animal reservoirs and human infections. Infection, Genetics and Evolution. 2014;21:523–530. doi: 10.1016/j.meegid.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 29.Paterson G. K., Larsen J., Harrison E. M., et al. First detection of livestock-associated meticillin-resistant Staphylococcus Aureus CC398 in bulk tank milk in the United Kingdom, January to July 2012. Eurosurveillance. 2012;17(50) [PMC free article] [PubMed] [Google Scholar]
- 30.Lozano C., López M., Gómez-Sanz E., Ruiz-Larrea F., Torres C., Zarazaga M. Detection of methicillin-resistant Staphylococcus aureus ST398 in food samples of animal origin in Spain. Journal of Antimicrobial Chemotherapy. 2009;64(6):1325–1326. doi: 10.1093/jac/dkp378. [DOI] [PubMed] [Google Scholar]
- 31.Podkowik M., Bystroń J., Bania J. Genotypes, antibiotic resistance, and virulence factors of staphylococci from ready-to-eat food. Foodborne Pathogens and Disease. 2012;9(1):91–93. doi: 10.1089/fpd.2011.0962. [DOI] [PubMed] [Google Scholar]
- 32.Balslev U., Bremmelgaard A., Svejgaard E., Havstreym J., Westh H. An outbreak of borderline oxacillin-resistant Staphylococcus aureus (BORSA) in a dermatological unit. Microbial Drug Resistance. 2005;11(1):78–81. doi: 10.1089/mdr.2005.11.78. [DOI] [PubMed] [Google Scholar]
- 33.Kernodle D. S., Classen D. C., Stratton C. W., Kaiser A. B. Association of borderline oxacillin-susceptible strains of Staphylococcus aureus with surgical wound infections. Journal of Clinical Microbiology. 1998;36(1):219–222. doi: 10.1128/jcm.36.1.219-222.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suggs A. H., Maranan M. C., Boyle-Vavra S., Daum R. S. Methicillin-resistant and borderline methicillin-resistant asymptomatic Staphylococcus aureus colonization in children without identifiable risk factors. The Pediatric Infectious Disease Journal. 1999;18(5):410–414. doi: 10.1097/00006454-199905000-00003. [DOI] [PubMed] [Google Scholar]
- 35.Perillo J., Ceccarelli D., Spagnoletti M., Lollai S., Cappuccinelli P., Colombo M. M. Molecular characterization of enterotoxigenic and borderline oxacillin resistant Staphylococcus strains from ovine milk. Food Microbiology. 2012;32(2):265–273. doi: 10.1016/j.fm.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 36.Kaszanyitzky É. J., Egyed Z., Jánosi S., et al. Staphylococci isolated from animals and food with phenotypically reduced susceptibility to β-lactamase-resistant β-lactam antibiotics. Acta Veterinaria Hungarica. 2004;52(1):7–17. doi: 10.1556/avet.52.2004.1.2. [DOI] [PubMed] [Google Scholar]
- 37.Cui S., Li J., Hu C., et al. Isolation and characterization of methicillin-resistant Staphylococcus aureus from swine and workers in China. Journal of Antimicrobial Chemotherapy. 2009;64(4):680–683. doi: 10.1093/jac/dkp275. [DOI] [PubMed] [Google Scholar]
- 38.Vestergaard M., Cavaco L. M., Sirichote P., et al. Sccmec type IX element in methicillin resistant Staphylococcus aureus spa type t337 (CC9) isolated from pigs and pork in Thailand. Frontiers in Microbiology. 2012;3, article 103 doi: 10.3389/fmicb.2012.00103.Article 103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Battisti A., Franco A., Merialdi G., et al. Heterogeneity among methicillin-resistant Staphylococcus aureus from Italian pig finishing holdings. Veterinary Microbiology. 2010;142(3-4):361–366. doi: 10.1016/j.vetmic.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 40.Mulders M. N., Haenen A. P. J., Geenen P. L., et al. Prevalence of livestock-associated MRSA in broiler flocks and risk factors for slaughterhouse personnel in the Netherlands. Epidemiology and Infection. 2010;138(5):743–755. doi: 10.1017/s0950268810000075. [DOI] [PubMed] [Google Scholar]
- 41.Rolo J., Miragaia M., Turlej-Rogacka A., et al. High genetic diversity among community-associated Staphylococcus aureus in Europe: results from a multicenter study. PLoS ONE. 2012;7(4) doi: 10.1371/journal.pone.0034768.e34768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Franco A., Hasman H., Iurescia M., et al. Molecular characterization of spa type t127, sequence type 1 methicillin-resistant Staphylococcus aureus from pigs. Journal of Antimicrobial Chemotherapy. 2011;66(6):1231–1235. doi: 10.1093/jac/dkr115. [DOI] [PubMed] [Google Scholar]
- 43.Sakwinska O., Giddey M., Moreillon M., Morisset D., Waldvogel A., Moreillon P. Staphylococcus aureus host range and human-bovine host shift. Applied and Environmental Microbiology. 2011;77(17):5908–5915. doi: 10.1128/aem.00238-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deurenberg R. H., Stobberingh E. E. The evolution of Staphylococcus aureus . Infection, Genetics and Evolution. 2008;8(6):747–763. doi: 10.1016/j.meegid.2008.07.007. [DOI] [PubMed] [Google Scholar]