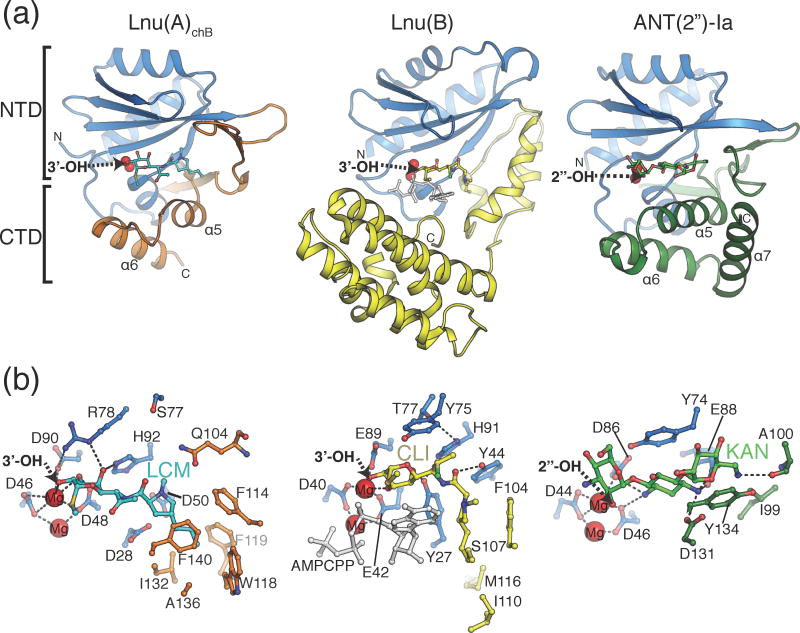
Fig. 5. Comparison of antibiotic recognition modes by Lnu(A), Lnu(B) and ANT(2″)-Ia.
A) Overall structures of Lnu(A)-LCM, Lnu(B)-CLI (PDB ID: 3JZ0) and ANT(2″)-Ia-KAN (PDB ID: 4WQK) complexes. The nucleotidyltransferase domains (NTD) are coloured light blue and the C-terminal domains (CTD) in orange, yellow and green, respectively, according to the enzyme. The antibiotic substrates are shown in sticks, the magnesium ions as red spheres and the nucleotidylation sites on the antibiotic molecules are labelled with arrows. The N- and C-termini are indicated with N and C, respectively.
B) Detailed comparison of enzyme-antibiotic interactions. Views are in the same orientation as Fig. 5A but zoomed in and amino acid sidechains that form hydrogen bonds or hydrophobic interactions with antibiotics are shown in sticks. For Lnu(B), the bound AMPCPP molecule is shown in ball-and-stick in light grey.