Figure 1. Identification of immunodominant and subdominant HLA-B*42:01-restricted epitopes.
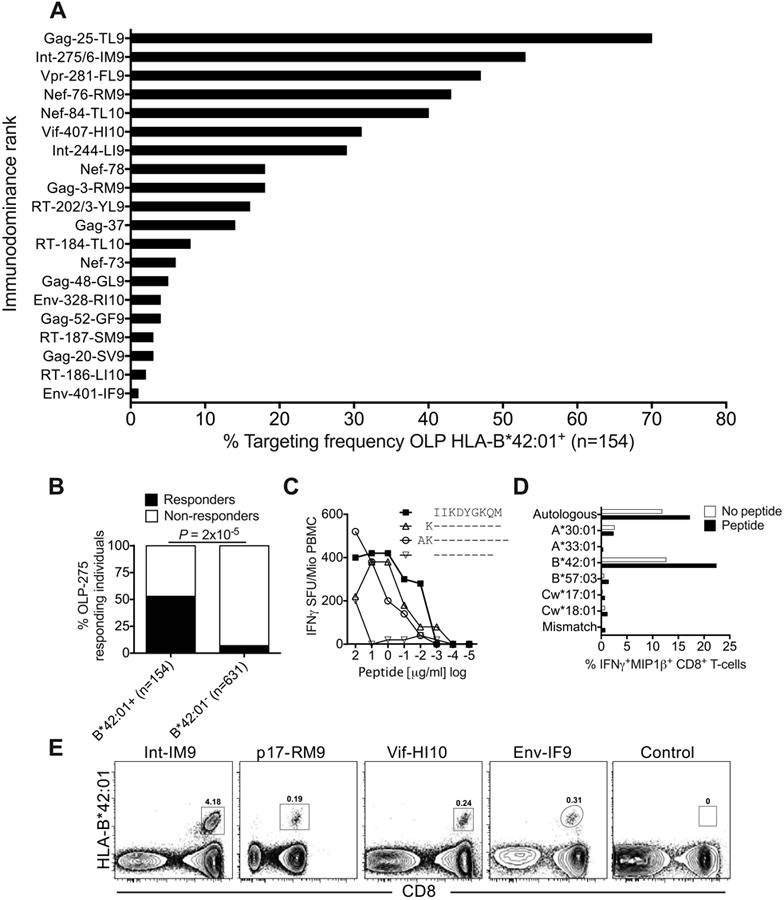
(A) Targeting frequencies for ‘protein-18mer peptide-optimal epitope name’ OLPs in HLA-B*42:01+ individuals, stratified for lack of HLA-B*07:02/39:10/42:02/81:01 co-expression (n=154). Responses were determined via IFNγ ELISpot assays. (B) Correlation between HLA-B*42:01 status and targeting of OLP-275 (KVVPRRKAKIIKDYGKQM) in IFNγ ELISpot assays. Significance was determined using Fisher's exact test. (C) Identification of the optimal HLA-B*42:01-restricted epitope Int-IM9 (IIKDYGKQM) via peptide truncations in IFNγ ELISpot assays. (D) Confirmation of HLA-B*42:01 as the restriction element for Int-IM9 via peptide pulsing of BLCLs partially HLA-matched to donor R014 (A*30:01/33:01, B*42:01/57:03, Cw17:01/18:01). Autologous or mismatched BLCLs were used as positive and negative controls, respectively. (E) Unequivocal confirmation of HLA-B*42:01 as the restriction element for Int-IM9 via cognate tetramer staining of responding PBMCs. Similar data are shown for three further novel epitopes: Gag-RM9 (RPGGKKHYM), Vif-HI10 (HPKVSSEVHI) and Env-IF9 (IPRRIRQGF). An HLA-mismatched tetramer was used as the negative control.
