Fig. 1.
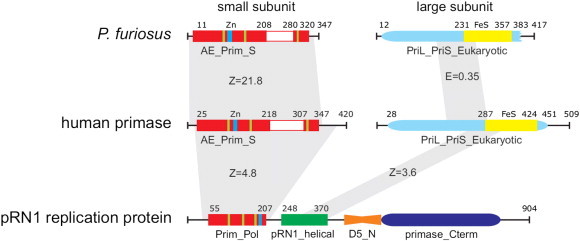
Domain organization and structural comparison of primases.
We compared the domain organization from Pyrococcus furiosus primase (small subunit: 1G71, large subunit: no structure available), human primase (4RR2) and the pRN1 multifunctional replication protein ORF904 (partial structure from amino acids 40–370, 3MIM). The domains of the respective proteins were determined with RPS-Blast and HHpred against the conserved domain database. Yellow rectangles indicate the position of the Fe–S cluster and cyan rectangles show the position of the zinc binding region. The white rectangles within the small primase subunits correspond to the unrelated helical domains which interrupt the prim fold. Green lines define the positions of three structurally highly conserved β-strands of the prim fold. The first β-strand harbors two conserved acidic residues, the second contains a highly conserved histidine and the last one is the flange running perpendicular to the other strands. Gray trapezoids highlight structural similarity as detected by DALI or BLAST if structural information is missing. The quality of the alignment is given with the Z-score or the E-value respectively. Numbers above each picture designate the limits of the domain borders and the length of the respective proteins. Proteins are drawn to scale.
