Fig. 6.
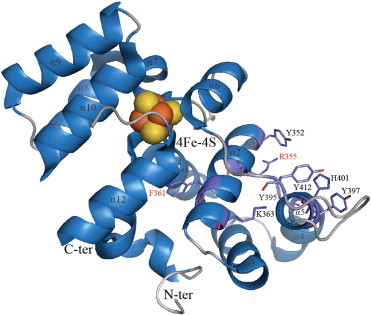
Putative DNA binding interface located on the C-ter part of the large primase subunit from S. cerevisiae.
The structural similarity between the PriL-CTD (3LGB) and the DNA-bound active site of DASH cryptochrome 3 from A. thaliana (2VTB) led Sauguet and collaborators [34] to propose a binding interface for the template. In this model, the large primase subunit (containing the FeS cluster) would assist the small one in the interaction with the DNA template. Key residues potentially involved in template binding are shown with purple, red and blue sticks corresponding to C, O and N atoms respectively. Most of them are located on helices 2 and 5. Invariant amino acids (see structure-based alignment in [34]) are highlighted in red. Orange and yellow spheres of the metal cluster (4Fe–4S) correspond to iron and sulfur atoms respectively. Residues 483 to 494 could not be detected.
