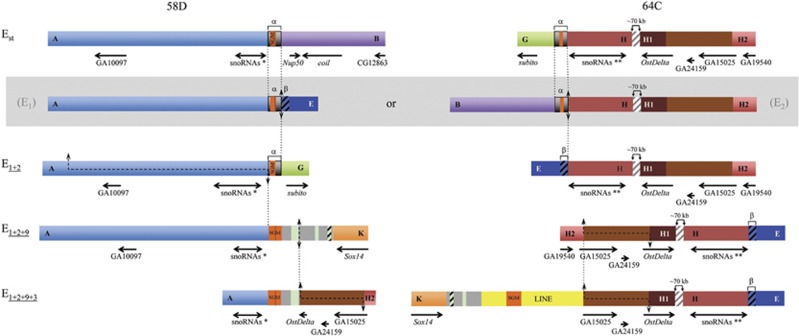
Figure 5.
Comparison of two reused breakpoints. The regions spanning the two breakpoints (at sections 58D and 64C, respectively) that have been multiply reused, at least at the cytological level (see Figure 1), in extant chromosomal arrangements Est, E1+2, E1+2+9 and E1+2+9+3 as well as in the two possible intermediate, and now extinct, arrangements connecting Est and E1+2 (within a gray box). Given that inversions occurred sequentially, vertical discontinuous lines connect a particular breakpoint in the corresponding non-inverted and inverted chromosomes. The blue striped fragments refer to a repeat motif, named β-motif in Puerma et al. (2014) and, therefore, marked with a β. The asterisk (*) and double asterisks (**) are meant to differentiate the snoRNAs generated from the Uhg5 and Uhg1 gene introns, respectively. The sequenced AB fragment is ~18.3-kb long. See legends of Figures 3 and 4 for all other notations.