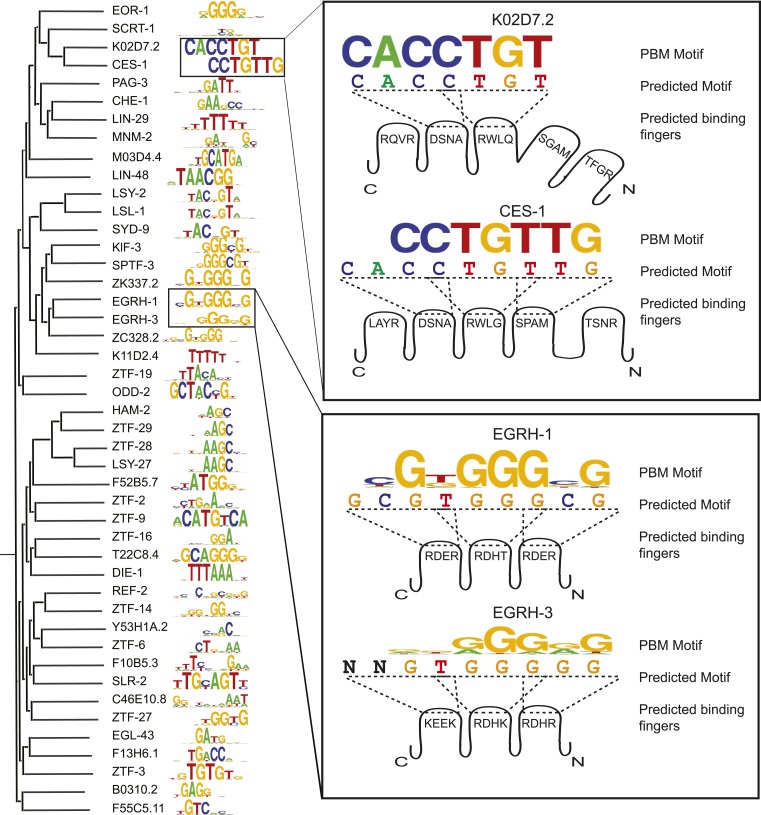
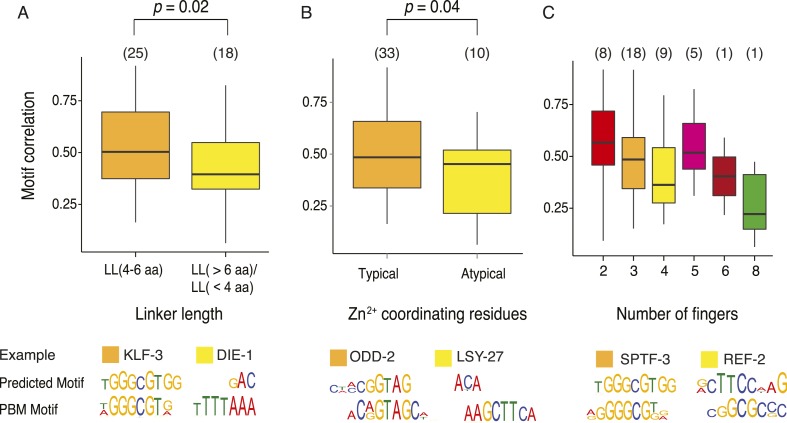
Figure 5. C2H2 motifs relate to DBD similarity and to the recognition code.
Left, ClustalW phylogram of C2H2 zinc finger (ZF) amino acid sequences with corresponding motifs. Right, examples in which motifs predicted by the ZF recognition code are compared to changes in DNA sequences preferred by paralogous C2H2 ZF TFs. Cartoon shows individual C2H2 ZFs and their specificity residues. Dashed lines correspond to 4-base subsites predicted from the recognition code.