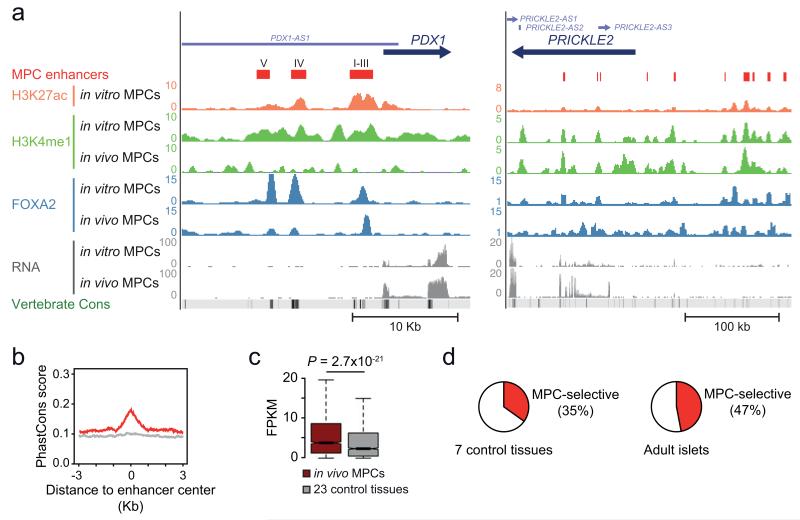
Figure 2.
A compendium of active enhancers in human pancreatic MPCs. (a) Predicted enhancers were defined by enrichment in H3K27ac and H3K4me1 (see schematic in Supplementary Fig. 2b). Shown are examples in the vicinity of PDX1, including a previously unannotated enhancer which we coin Area V, upstream of known enhancers (Areas I-IV)42,43, and several enhancers near PRICKLE2, a non-canonical WNT signaling component (Supplementary Table 4). (b) MPC enhancer sequences are evolutionary conserved (17 species vertebrate PhastCons score). Conservation plots of random non-exonic sequences are shown as a light gray line. (c) Genes that are associated with 3 or more MPC enhancers show enriched expression in dissected in vivo MPCs relative to 23 other tissues. The boxes show interquartile range (IQR) of RNA levels, whiskers extend to 1.5 times the IQR or extreme values, and notches indicate 95% confidence intervals of the median. P value was calculated with Wilcoxon rank-sum test (n=2,093 genes). (d) Many MPC enhancers are tissue- and stage-selective. We defined enhancers of 8 control tissues using identical criteria as in MPCs (Supplementary Fig. 2c, Supplementary Table 8) and show the proportion of enhancers that are inactive in at least 6 out of 7 non-pancreatic tissues (left) or inactive in adult pancreatic islets (right).