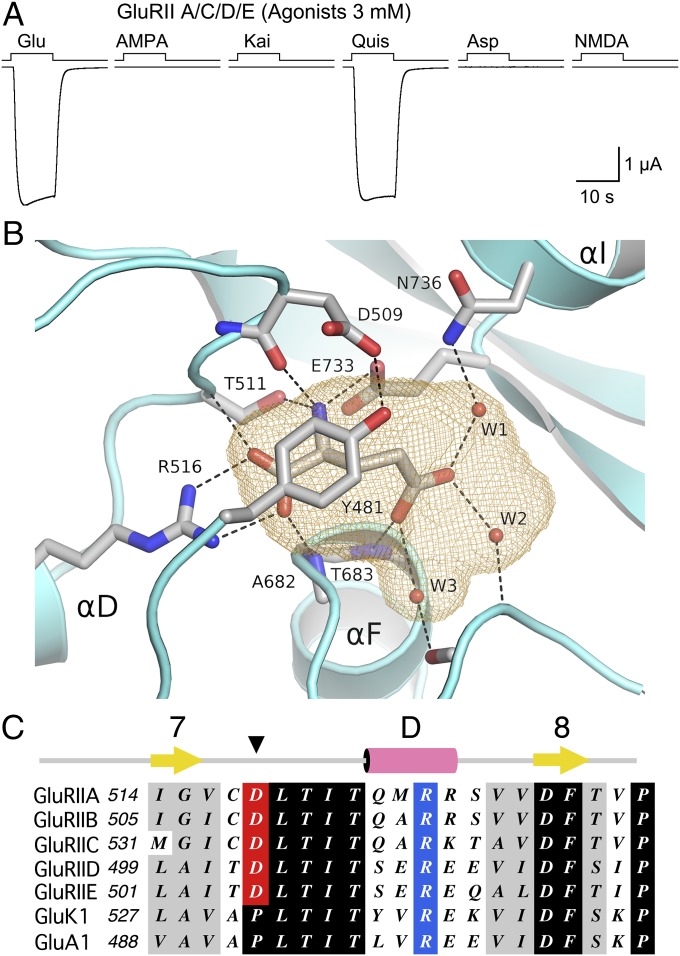
Fig. 5.
Structural basis for ligand selectivity of Drosophila NMJ iGluRs. (A) Responses at −60 mV to sequential applications of six glutamate receptor agonists, all at 3 mM, recorded from a single oocyte expressing Neto β and GluRIIA/C/D/E. (B) Ribbon diagram of the GluRIIB ligand binding domain crystal structure showing a bound glutamate molecule trapped in a cavity (orange mesh) together with three water molecules. The cavity is capped by Tyr481 which is held in place by a hydrogen bond formed with the side chain of Asp509. (C) Sequence alignments for part of domain 1 for five Drosophila NMJ iGluRs, and representative vertebrate kainate (GluK1) and AMPA (GluA1) receptors, showing the exchange of a conserved proline residue for an aspartate in the Drosophila NMJ iGluRs.